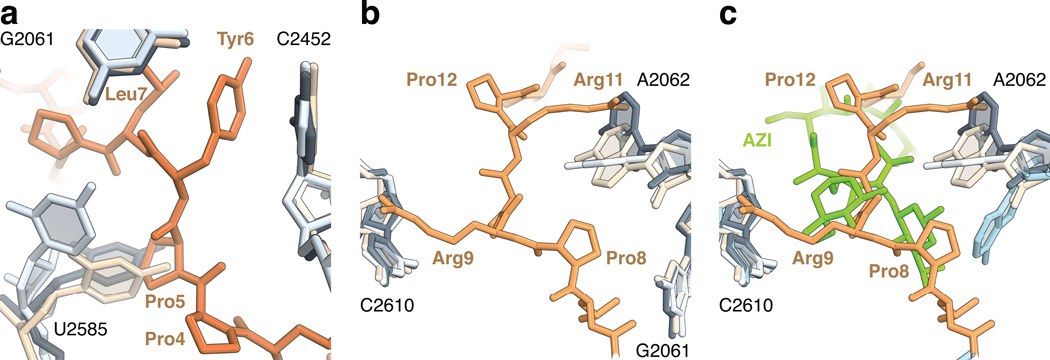
Figure 3. Conformational changes in the peptide exit tunnel.
(a) In the complex of onc112 with the 70S ribosome, the universally conserved nucleotide U2585 (2596) (light blue) changes its position to accommodate Pro5 (70S-CAM complex, ivory, PDB accession 4V7W 18; 70S complex with A- and P-site tRNAs, dark grey, PDB accession 1VY4 17). (b) In the upper chamber of the peptide exit tunnel, the binding of onc112 fixes the conformation of the nucleotide base of A2062 (2083) (light blue) such that it forms a stacking interaction with Arg11 and accommodates Pro8. (c) Same as in panel b, but showing the macrolide antibiotic azithromycin (AZI, green) and A2062 (blue) from the structure of the 70S-AZI complex (PDB accession 4V7Y) 18.