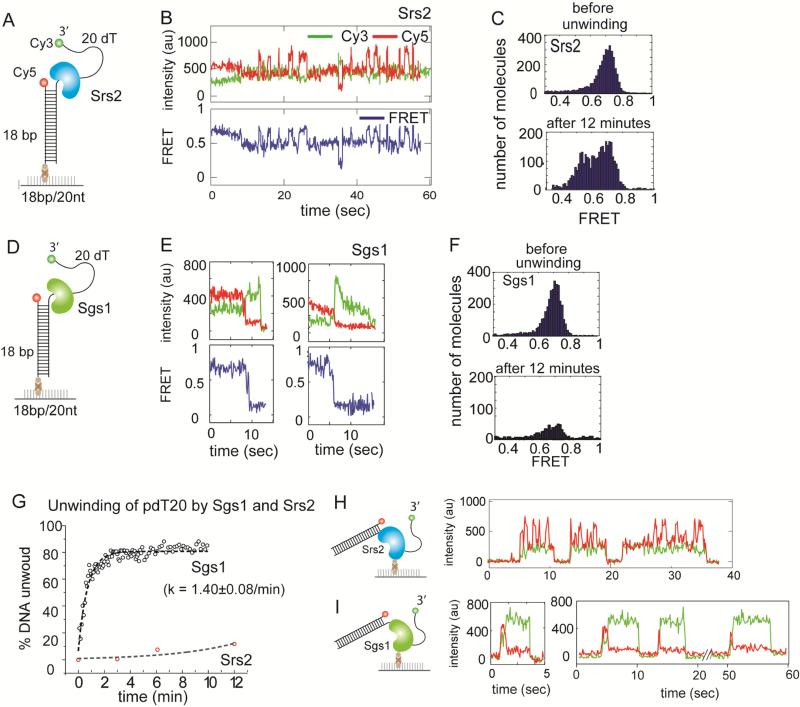
FIGURE 1. Unwinding of duplex DNA by Srs2 and Sgs1, also see Figure S1.
(A, D) Schematic of partial duplex DNA with donor (Cy3) and acceptor (Cy5) fluorophores used for unwinding. (B, E) Representative single molecule intensity (top) and FRET (bottom) traces obtained from applying 10nM Srs2 or Sgs1 to the unwinding substrate, respectively. (C) FRET histogram taken before and after unwinding reaction by Srs2 showing no unwinding. (F) FRET histogram taken before and after unwinding reaction by Sgs1 showing rapid unwinding. (G) Unwinding rate of Sgs1 (black circle) and Srs2 (red circle) and the first exponential fit. Data are represented as mean ± S.E.M.. (H, I) Histidine-tagged Srs2 or flag-tagged Sgs1 proteins (0.5-1nM) were each immobilized on a surface treated with biotinylated Ni–NTA (nitroloacetic acid) or biotinylated anti-flag antibody, respectively and the representative single molecule traces obtained in each case are shown.