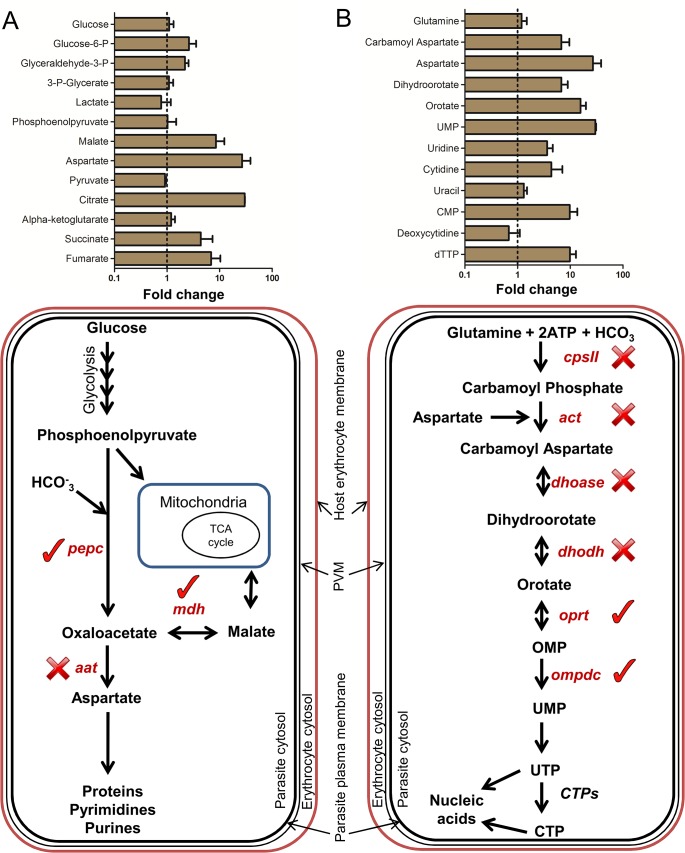
Fig 2. Metabolites of intermediary carbon metabolism (ICM) and pyrimidine biosynthesis are up-regulated in reticulocytes.
A. Top panel: Fold change of relative levels (peak intensities) of metabolites of carbon metabolism in rodent REP compared to wtEP. Dotted line indicates no change and error bars indicate R.S.D. (Relative Standard Deviation) of peak intensities from reticulocyte samples multiplied to the fold change values from n = 3 independent biological replicates. Bottom panel: Schematic representation of intermediary carbon metabolism (ICM) in Plasmodium cytosol. Genes marked with (✓) were deleted in P. berghei blood stages and the ones marked with (✕) could not be deleted even after repeated attempts. pepc: Phosphoenolpyruvate Carboxylase (PBANKA_101790), mdh: Malate Dehydrogenase (PBANKA_111770), aat: Aspartate Amino Transferase (PBANKA_030230). B. Top panel: Fold change of relative levels (peak intensities) of metabolites of pyrimidine biosynthesis in rodent REP compared to wtEP. Dotted line indicates no change and error bars indicate R.S.D. (Relative Standard Deviation) of peak intensities from reticulocyte samples multiplied to the fold change values from n = 3 independent biological replicates. Bottom panel: Schematic representation of pyrimidine biosynthesis pathway in Plasmodium cytosol. Genes marked with (✓) were deleted in P. berghei blood stages and the ones marked with (✕) could not be deleted even after repeated attempts. cpsII: Carbamoyl phosphate synthetase II (PBANKA_140670), act: Aspartate carbamoyltransferase (PBANKA_135770), dhoase: Dihydroorotase (PBANKA_133610), dhodh: Dihydroorotate dehydrogenase (PBANKA_010210), oprt: Orotate phosphoribosyltransferase (PBANKA_111240), ompdc: Orotidine 5′-monophosphate decarboxylase (PBANKA_050740). (Also see Fig B in S1 Text for gene deletion strategy and confirmation)