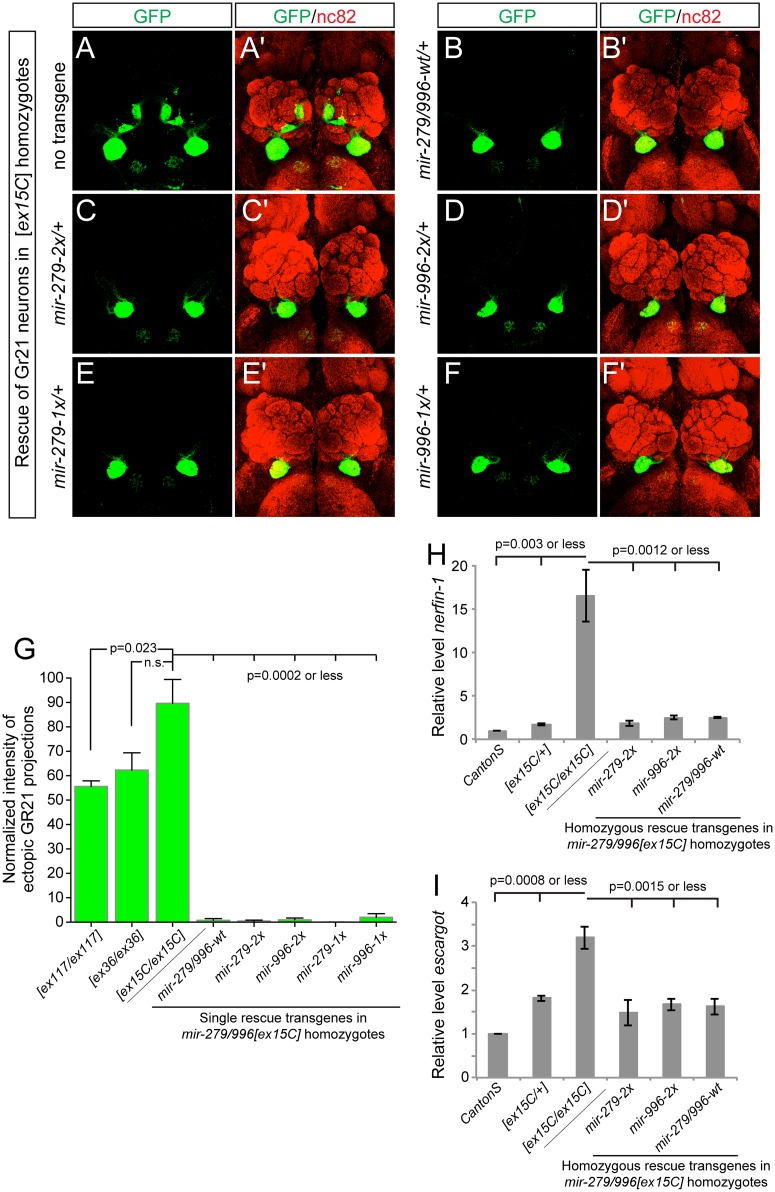
Fig 4. miR-996 restores proper CO2-sensing neurons to mir-279/996 double mutants.
(A-F) Representative images of GR21 projections in brains from mir-279/996[ex15C] whole animal mutants and rescued backgrounds bearing one copy of each 16.6 kb genomic transgene. Genotype for the mutant is: Gr21-Gal4, UAS-syt-GFP/+; mir-279/996[ex15C], and rescued genotypes are: Gr21-Gal4, UAS-syt-GFP/mir[rescue]; mir-279/996[ex15C]. (A, A') Double deletion mir-279/996[ex15C] homozygote. (B-F) Rescue of mir-279/996[ex15C] homozygotes by a copy of the wildtype transgene (B, B'), by a single mir-279-only transgene (C, C'), by a single mir-996-only transgene (D, D'), by a two-copy mir-279-only transgene (E, E'), and by a two-copy mir-996-only transgene (F, F'). (G) Quantification of ectopic Gr21 axon projection normalized to the normal ventral projection. 4–6 brains were analyzed for each genotype, error bars represent SEM, and p-values calculated by Student's two-tailed T-test, equal variance. (H-I) qPCR assays show elevated nerfin-1 (H) and escargot (I) transcript levels in mir-279/996[ex15C] homozygous heads. These defects are comparably rescued by mir-279-2x or mir-996-2x transgenes as they are by wildtype mir-279/996 genomic transgene, to target levels that are slightly higher than in Canton S heads but similar to [ex15C]/+ heads. Error bars represent SD, p-values calculated using unpaired T-tests from the combined data of independent, triplicate qPCR assays.