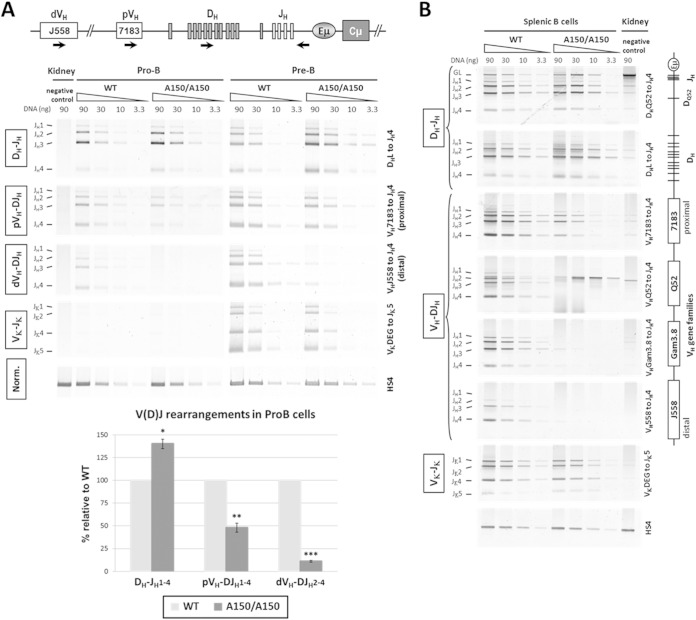
FIG 2.
V(D)J rearrangement analyses. (A) Genomic DNAs were prepared from sorted pro-B and pre-B cells and were subjected to semiquantitative PCR to amplify D-JH, VH-DJH, and Vκ-Jκ rearrangements, using primers that bind the indicated segments and primers that pair 3′ of JH4 (for the IgH locus) or 3′ of Jκ5 (for the κ locus). Kidney DNA was used as a negative control. A PCR analysis of the HS4 enhancer from the 3′RR was used for normalization of DNA input. The PCR for VκJκ rearrangement also allowed us to check the purity of the sorted populations (n = 3 for each experiment, with a pool of >5 mice). Quantification of the recombination events at the IgH locus of pro-B cells is displayed in the histogram. (B) Genomic DNAs were prepared from sorted splenic B cells and subjected to PCR as described for panel A (n = 2). The relative positions of the VH gene families are indicated on the right. The signals detected for the VHQ52 gene segment in the A150 lanes were derived from the inserted VH gene. Data are presented as means ± SEM. ***, P < 0.001; **, P < 0.01; *, P < 0.05.