Figure 9.
Retromer Is Required for Autophagy Processes.
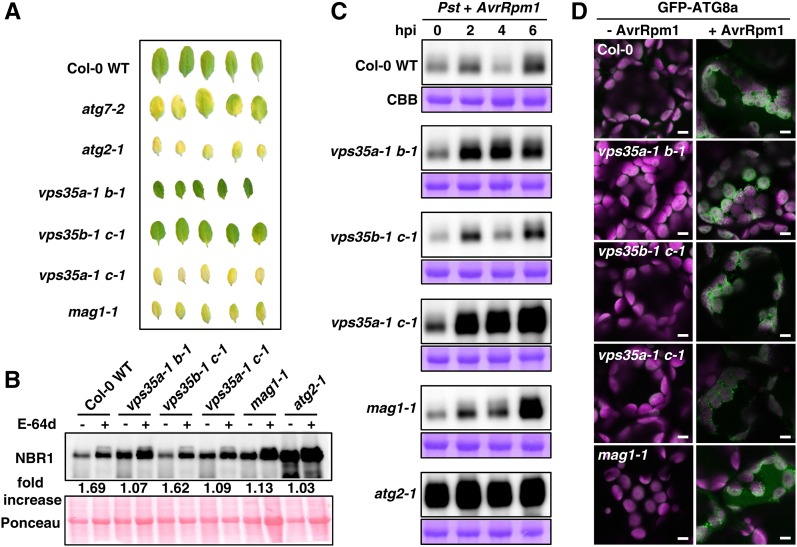
(A) Leaf detachment assay of Col-0 wild-type, atg7-2, and atg2-1 plants compared with vps35a-1 b-1, vps35a-1 c-1, and vps35b-1 c-1 double mutants and mag1-1. Detached leaves of 4-week-old plants were kept for 4 d on moist filter paper in darkness.
(B) Immunoblot analysis of NBR1 accumulation in vps35a-1 b-1, vps35b-1 c-1, vps35a-1 c-1, and mag1-1 mutant seedlings grown for 10 d on MS medium before treatment with DMSO (−) or the cysteine protease inhibitor E-64d (+) for 12 h. Equal amounts of crude extracts were separated by SDS-PAGE and probed on blots with anti-NBR1 antibody. Numbers correspond to the fold increase of NBR1 in E-64d-treated samples compared with the respective DMSO control. Ponceau staining of membrane-bound total proteins was used as a loading control and quantified for the normalization of NBR1 signal intensities.
(C) Immunoblot analysis of NBR1 accumulation upon infection with Pst DC3000 (AvrRpm1). Total proteins were extracted at the indicated time points from 5-week-old retromer mutants compared with Col-0 wild-type and atg2-1 controls and probed with anti-NBR1 antibody (top). Bottom panels indicate Coomassie Brilliant Blue (CBB) staining of Rubisco large subunit as a loading control.
(D) GFP-ATG8a accumulation is indicative of autophagosome formation in the Col-0 wild-type and retromer mutants upon infection with Pst DC3000 (AvrRpm1). Confocal images are representative of palisade parenchyma cells in leaves of 4-week-old plants before (−AvrRpm1) and 3 h after (+AvrRpm1) infection. GFP-ATG8a derived signals (green) are superimposed with chlorophyll fluorescence of chloroplasts (magenta). Bars = 5 μm.