Figure 1.
Analyses of Gle1-Silencing Phenotypes Using DEX-Inducible RNAi in Arabidopsis.
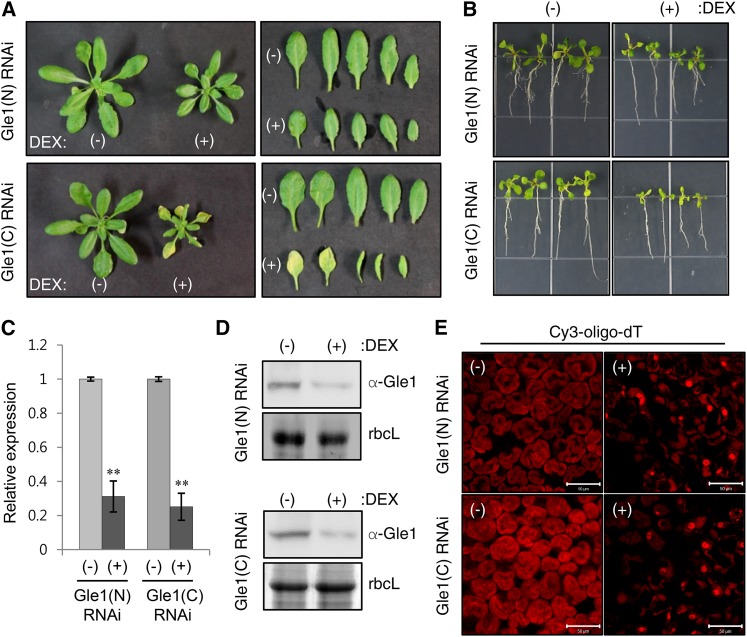
(A) Plant phenotypes of two different Arabidopsis DEX-inducible Gle1 RNAi lines [Gle1(N) and Gle1(C) RNAi] upon DEX treatment. Plants were grown in soil for 14 d and then sprayed with either ethanol (−) or 30 μM DEX (+) for 7 d.
(B) Seedling phenotypes of the two Gle1 RNAi lines that were grown for 10 d on MS medium containing either ethanol (−) or 10 μM DEX (+).
(C) Real-time quantitative RT-PCR analysis to determine Gle1 transcript levels. Transcript levels in (+)DEX samples are expressed relative to those in (−)DEX samples. UBC10 mRNA levels were used as a control. Values represent means ± sd of three replicates per experiment. Asterisks denote statistical significance of the differences between (−)DEX and (+)DEX samples based on two-tailed Student’s t tests: *P ≤ 0.05 and **P ≤ 0.01.
(D) Immunoblotting with anti-Gle1 antibodies to determine the endogenous Gle1 protein levels. Coomassie blue-stained rbcL (Rubisco large subunit) was used as a control.
(E) In situ hybridization of leaves of the two Gle1 RNAi lines after spraying with ethanol (−) or 30 μM DEX (+) for 7 d using the Cy3-oligo-dT probe for confocal microscopy. Bars = 50 μm.