Fig. 2.
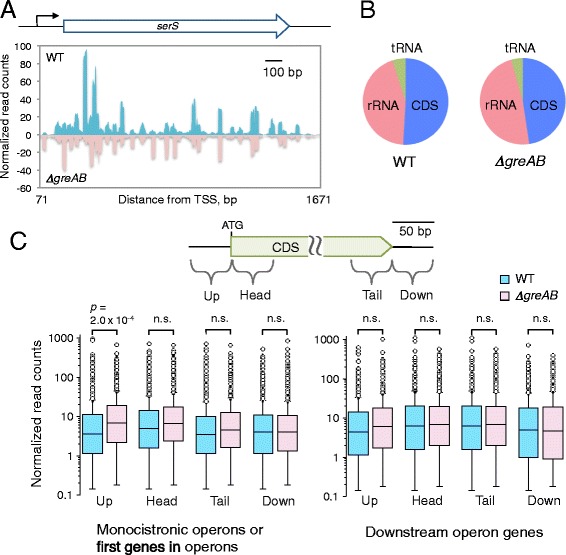
Comparison of genome-wide transcription in E. coli WT and ΔgreAB cells. (a) A transcription pausing profile of the serR gene. TSS transcription start site. (b) Mapped sequencing reads from paused RNAP complexes carrying mRNA (coding DNA sequence (CDS)), tRNA and rRNA. (c) GreAB proteins reduce pausing in 5′ UTRs of E. coli mRNA genes. Each box plot represents the quartile of normalized read counts in a 50-bp window for each gene body: upstream (Up), head, tail, and downstream (Down). mRNA genes with normalized read counts >0.1 (n = 1847 for left panel and n = 882 for right panel) were used for the analysis. The p-value of two-tailed t-test is shown for a pair with statistically significant difference between the WT and ΔgreAB data. The p-values >0.05 are labeled as non-significant (n.s.)
