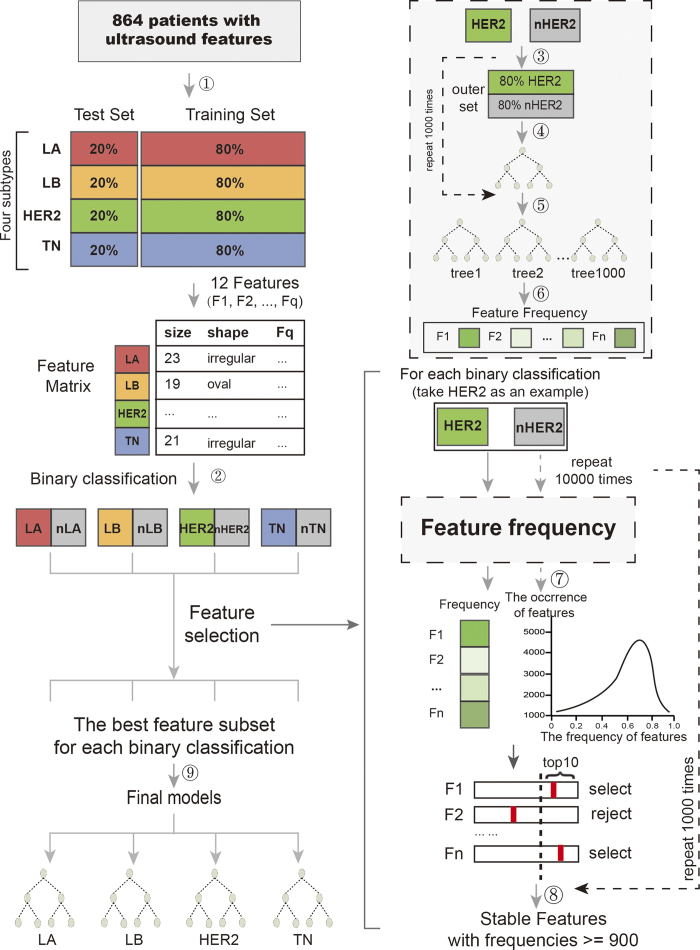
Figure 7. The step-by-step protocol for the ensemble approach computational algorithm.
① Construct the training and test set by sampling patients. The training set and test set contain 80% and 20% of the initial patients for each subtype, respectively. ② Binary classification: LA, LB and TN are classified as one category, called nHER2 (take HER2 as an example). ③ Randomly sample patients, 80% HER2 and 80% nHER2, from the training set, called the outer set. ④ Grow a tree on the particular set obtained from step 2. ⑤ After repeating steps 2-4 1,000 times, we obtained 1,000 trees. ⑥ Count the occurrence frequencies of all features in the above 1,000 trees. ⑦ Randomly assign a label to each patient. Then, use the shuffled data to conduct steps 1-5. Compute the relevance intensity, (FV) 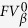 , for each feature. Repeat the permutation 10,000 times to obtain an empirical threshold for the specified significance level of β. ⑧ After repeating steps 2–6 1,000 times, we obtained the stable high occurrence frequency of features. ⑨ Construct the model based on the selected features.
, for each feature. Repeat the permutation 10,000 times to obtain an empirical threshold for the specified significance level of β. ⑧ After repeating steps 2–6 1,000 times, we obtained the stable high occurrence frequency of features. ⑨ Construct the model based on the selected features.