Figure 1.
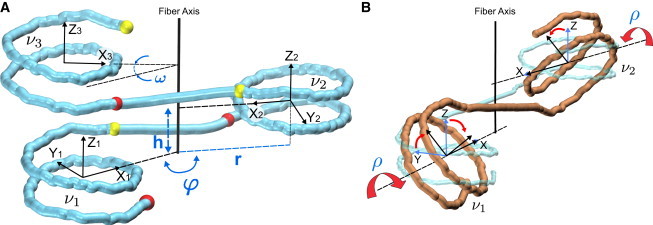
Definition of the chromatin fiber parameters. (A) Cylindrical parameters h (rise), r (radius), and polar angle φ. This angle determines the superhelical fiber twist: ω = 2φ –360° (the polar angle between the centers of nucleosomes ν1 and ν3). In this particular case, φ = 165° and ω = –30°. The entry points of nucleosomes are shown as red balls, and the exit points as yellow balls. Each nucleosome is associated with a right-handed coordinate frame in which axis Z represents the superhelical axis of the nucleosome (calculated as described in (44)), axis X points toward the nucleosome dyad, and axis Y is perpendicular to X and Z. (B) Inclination angle ρ (defining rotation around the dyad axis X) is related to the angle γ measured by electric dichroism (39–41), which is the angle between the Y axis and the fiber axis. These two angles are related by equation γ = | 90° – | ρ | |. Increasing the inclination angle by Δρ leads to decreasing the total twisting of the linker between nucleosomes ν1 and ν2 by ∼2Δρ. The images were prepared with the VMD software package (45). To see this figure in color, go online.
