Figure 8.
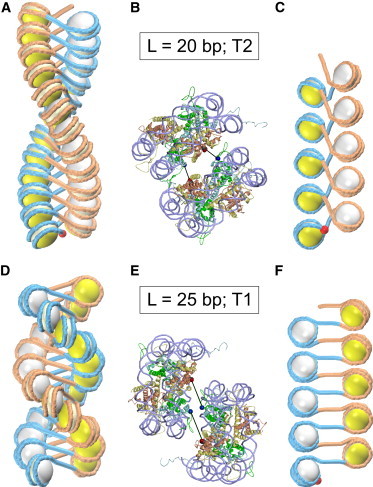
Topoisomers T2 and T1 for linkers L = 20 bp (A–C) and L = 25 bp (D–F), respectively. The optimal structure for L = 20 bp has inclination angle ρ = –45° and rise h = 27 Å (A and B); the optimal structure for L = 25 bp has ρ = 90° and h = 23 Å (D and E). The difference between the two families is clearly visible in the stretched forms with h = 55 Å (C and F). Note a more extensive folding of DNA in the nucleosomal disks in (C). This is consistent with the DNA writhing: Wr = –1.7 in (C) and Wr = –0.9 in (F). Red balls indicate the DNA entry points. The detailed representations in (B) and (E) show that there are no steric clashes between the stacked nucleosomes ν1 and ν3 (Fig. 1A). The internucleosome stacking is stabilized by the H4 tail-acidic patch bridges shown by black lines; the distances between Asp24 (H4) and Glu61 (H2A) are 23 Å in (B) and 34 Å in (E) (see Fig. S3 for details). To see this figure in color, go online.
