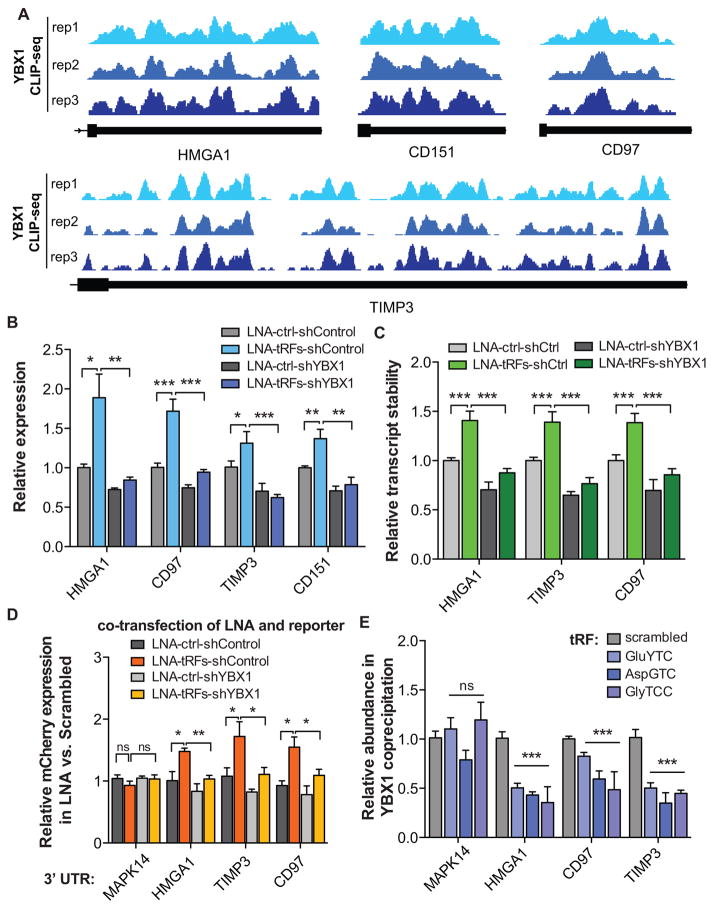
Figure 5. YBX1 target transcripts and their response to changes in tRF levels.
(A) YBX1 interacts with the 3′UTRs of HMGA1, CD151, CD97, and TIMP3. The last exon of the indicated transcripts are shown with mapped reads from experimental replicates of YBX1 CLIP-seq. (B) Transfection of antisense LNAs against YBX1-binding tRFs resulted in the up-regulation of HMGA1, CD151, CD97, and TIMP3 transcripts, in a YBX1-dependent manner, as determined by qPCR measurements. (C) Similarly, transfecting antisense LNAs resulted in a significant stabilization of HMGA1, CD97, and TIMP3 transcripts in a YBX1-dependent manner. Whole-genome RNA stability measurements were perfomed using α-amanitin-mediated inhibition of RNA polymerase II (see Methods). (D) A GFP/mCherry dual-reporter assay was used to measure the effects of cloning HMGA1, CD97, and part of the TIMP3 3′ UTRs downstream of mCherry using qRT-PCR. The 3′ UTR of MAPK14, which is devoid of YBX1 tags, was included as a control. Consistent with our prior findings, LNA transfections resulted in a significant increase in relative mCherry expression in a YBX1-dependent manner. (E) Exogenously added tRF mimetics, while showing no effect on MAPK14 abundance, resulted in a significant depletion of HMGA1, CD97, and TIMP3 transcripts from the YBX1 co-immunoprecipitated RNA population. Statistical significance is measured using one-tailed Student’s t-test: *, p<0.05, **, p<0.01, and ***, p<0.001. Error bars in all panels indicate s.e.m. unless otherwise specified.