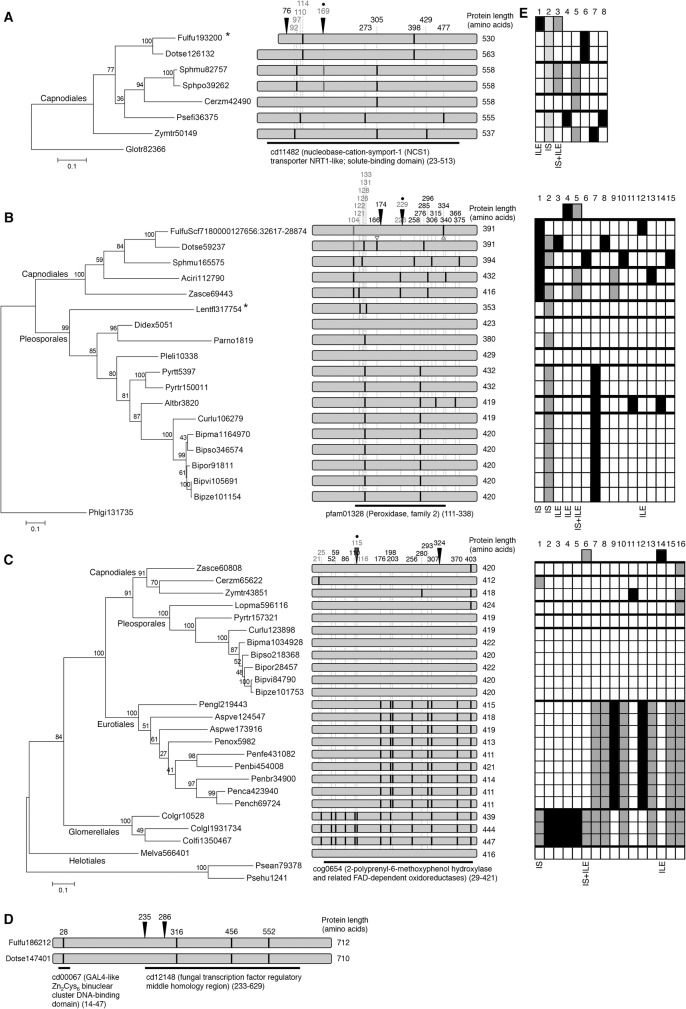
Fig 3. Analysis of intron positions in genes with recent insertions of Introner-Like Elements (ILE).
ILEs from Passalora brachycarpa inserted in genes encoding (A) a transporter and (B) a peroxidase. ILEs from Passalora miurae inserted in genes encoding (C) a hydroxylase/oxidoreductase and (D) a fungal transcription factor. For each gene, a maximum likelihood phylogenetic tree was constructed with the predicted protein sequence of orthologues. The trees were rooted with the closest homologue found in Basidiomycota. Bootstrap values of 100 repeats are shown. Scale bar represents the number of substitutions per site. The numbers correspond to the protein ID from the Joint Genome Institute mycocosm portal, except for one gene that was not predicted in the Fulvia fulva genome and for which genomic coordinates are given (B). Orders in fungal classification are mentioned in the trees. On the right, diagrams depict aligned protein sequences and intron positions are indicated as black bars. Their positions in the protein alignment that served to build the phylogenetic trees are indicated above. Positions that are shown in grey highlight putative intron sliding. The black arrows indicate the positions where ILEs inserted in the genes of P. miurae or P. brachycarpa. The open triangles indicate previously identified ILEs. Dots indicate positions where parallel intron gains have occurred. The black bar below each protein representation indicates conserved domains (positions in the protein alignments are indicated between brackets). Asterisks behind species names indicate genes that are likely pseudogenes because of an in frame stop codon. (E) Schematic overview of intron positions (numbered on top) in three genes. The first row shows intron positions in genes of P. miurae or P. brachycarpa. Thick lines indicate monophyletic clades according to the phylogenetic trees. Black, dark grey and light grey squares indicate single presence of an intron position in a monophyletic clade, presence-absence polymorphism and single absence of an intron position in a monophyletic clade, respectively. White squares indicate absence of intron. The presence of ILE and occurrence of putative intron splicing (IS) are indicated below each scheme. Aciri: Acidomyces richmondensis; Altbr: Alternaria brassicicola; Cerzm: Cercospora zeae-maydis; Fulfu: Fulvia fulva; Bipze: Bipolaris zeicola; Bipma: Bipolaris maydis; Curlu: Curvularia lunata; Bipor: Bipolaris oryzae; Bipso: Bipolaris sorokiniana; Bipvi: Bipolaris victoriae; Didex: Didymella exigua; Dotse: Dothistroma septosporum; Lentfl: Lentithecium fluviatile; Pleli: Plenodomus lingam; Lopma: Lophiostoma macrostomum; Psefi: Pseudocercospora fijiensis; Pyrtr: Pyrenophora tritici-repentis; Pyrtt: Pyrenophora teres f. teres; Sphmu: Sphaerulina musiva; Sphpo: Sphaerulina populicola; Parno: Parastagonospora nodorum; Zasce: Zasmidium cellare; Zymtr: Zymoseptoria tritici; Aspve: Aspergillus versicolor; Aspwe: Aspergillus wentii; Penbi: Penicillium bilaiae; Penbr: Penicillium brevicompactum; Penca: Penicillium canescens; Pench: Penicillium chrysogenum; Penfe: Penicillium fellutanum; Pengl: Penicillium glabrum; Penox: Penicillium oxalicum; Colgr: Colletotrichum graminicola; Colfi: Colletotrichum fiorinae; Colgl: Colletotrichum gloeosporioides; Melva: Meliniomyces variabilis; Glotr: Gloeophyllum trabeum; Phlgi: Phlebiopsis gigantean; Psean: Pseudozyma antarctica; Psehu: Pseudozyma hubeiensis.