Abstract
In hot deserts, plants cope with aridity, high temperatures, and nutrient-poor soils with morphological and biochemical adaptations that encompass intimate microbial symbioses. Whereas the root microbiomes of arid-land plants have received increasing attention, factors influencing assemblages of symbionts in above-ground tissues have not been evaluated for many woody plants that flourish in desert environments. We evaluated the diversity, host affiliations, and distributions of endophytic fungi associated with photosynthetic tissues of desert trees and shrubs, focusing on non-succulent woody plants in the species-rich Sonoran Desert. To inform our strength of inference, we evaluated the effects of two different nutrient media, incubation temperatures, and collection seasons on the apparent structure of endophyte assemblages. Analysis of >22,000 tissue segments revealed that endophytes were isolated four times more frequently from photosynthetic stems than leaves. Isolation frequency was lower than expected given the latitude of the study region, and varied among species a function of sampling site and abiotic factors. However, endophytes were very species-rich and phylogenetically diverse, consistent with less-arid sites of a similar latitudinal position. Community composition differed among host species, but not as a function of tissue type, sampling site, sampling month, or exposure. Estimates of abundance, diversity and composition were not influenced by isolation medium or incubation temperature. Phylogenetic analyses of the most commonly isolated genus (Preussia) revealed multiple evolutionary origins of desert-plant endophytism and little phylogenetic structure with regard to seasonality, tissue preference, or optimal temperatures and nutrients for growth in vitro. Together, these results provide insight into endophytic symbioses in desert plant communities, and can be used to optimize strategies for capturing endophyte biodiversity at regional scales.
Keywords: Arid lands, Ascomycota, diversity, Dothideomycetes, fungi, Larrea, phylogeny, Parkinsonia, Preussia, Simmondsia, symbiosis
Introduction
One of the most profound insights of the past century is that no macroscopic organism exists in nature in the absence of microbes [60]. From the bacterial microflora associated with the digestive systems of animals to the mycorrhizal fungi that enhance nutrient uptake and water-use efficiency in most extant plants, microorganisms play critical but often overlooked roles in the biological interrelationships that underlie all ecosystems [43, 61, 89].
In hot deserts such as those of the American southwest, plants cope with aridity, high temperatures, intense solar radiation, nutrient-poor soils, and other physiological and ecological challenges with well-documented morphological and biochemical adaptations [62, 90]. Less well explored, however, are the intimate associations of such plants with microbes, which may confer thermotolerance, drought resistance, and other important benefits that enhance survival, primary productivity, and plant community structure [58, 91].
Recently, root-symbiotic fungi such as mycorrhizae and dark-septate endophytes have received increasing attention from biologists in arid lands [1, 15, 18, 31, 34, 47, 48, 52, 53, 55, 59, 67, 68, 92]. Such work has greatly enhanced our understanding of the geographic-, host-, and tissue-affiliations of fungi that are symbiotic with ecologically important plants in semi-arid grasslands and deserts, especially grasses and sedges [34, 47], gypsophilous plants [69], and selected dicotyledonous plants such as Atriplex spp. [16, 54] and some Cactaceae [17, 83; see also 51]. However, gaps remain with regard to understanding the diversity and composition of fungal communities in above-ground tissues of many woody plants in arid ecosystems, including those non-succulent species that form the dominant cover in some of the world's most biotically rich deserts. The Sonoran Desert, which spans ca. 280,000 km2 in the southwestern USA and northern Mexico, is the hottest and most biotically rich desert in North America, but little is known about the endophyte communities associated with its most prevalent plant species (but see [9, 11, 83]).
Fungal endophytes are microscopic fungi that occur within apparently healthy tissues such as roots and shoots of living plants [74]. Known from every plant species examined to date, endophytes form symbioses with plants in terrestrial communities ranging from tundra to tropical rainforests, agricultural systems, and hot deserts [5, 6, 9, 11, 22, 30, 34, 38, 65, 69, 75, 83]. Although long overlooked because of their cryptic occurrence in symptomless tissues, endophytes are increasingly recognized for their ecological importance, especially in extreme environments [75]. Endophytes in above-ground tissues such as leaves and stems, which are horizontally transmitted and typically form highly localized infections, represent one of many functional groups of endophytes [74]. These endophytes – characterized in [74] as class 3 endophytes – are the focus of this study.
Class 3 endophytes (hereafter, endophytes) that associate with photosynthetic tissues of most plants often are highly diverse at the scales of individual plants, plant communities, and biogeographic regions [22, 28, 86]. The few species studied in detail thus far influence hosts' tolerance of heat stress, drought, soil salinity, herbivory, and disease, suggesting both short-term and evolutionary impacts on plants' physiological and ecological traits [6, 10, 13, 30, 65, 73-75]. These fungi produce a diversity of metabolites of interest in medicinal drug discovery, biofuels development, bioremediation, industrial applications, and biological control [32, 35, 76, 80, 81]. Thus, understanding their distributions is important both for illuminating features of plant ecology and evolution in natural and human-made systems, and for designing effective survey strategies for tapping their biochemical resources [35, 77].
Recent studies of leaf- and stem endophytes in woody plants have suggested a latitudinal gradient of abundance, diversity, and host specificity [9, 11]: in culture-based studies, abundance and species richness are especially high in tropical forests, but phylogenetic diversity and host specificity tend to be more pronounced at higher latitudes. In turn, factors such as annual precipitation, plant density, land use history, and related variables often are associated with distinctive endophyte communities [8, 27, 37, 50, 87], potentially providing exceptions to the latitudinal gradients observed thus far. The spatial scale at which these endophytes turn over in community structure also varies among biomes and as a function of climate [23, 37, 50], but little information is available for endophytes in woody plants in desert ecosystems.
Although important under many conditions, symbioses between plants and fungi are thought to manifest their greatest importance in stressful environments [30, 73, 74], where close associations with hosts may mitigate or overcome particular abiotic or biotic challenges. Studies of endophytes in arid lands that are home to phylogenetically diverse plant communities, but that also constitute relatively extreme conditions, provide an opportunity to evaluate the contributions of previously unknown endophyte communities to global estimates of diversity and broader perspectives in endophyte-plant interactions.
The goal of this study was to evaluate the abundance, diversity, tissue- and host affiliations, and local distributions of endophytic fungi associated with photosynthetic tissues of desert trees and shrubs. We focused on leaves and stems of the most common non-succulent woody plants in the Arizona Upland subdivision of the Sonoran Desert bioregion, with focal plants located in diverse microhabitats in a large area of a contiguous, protected, and biotically consistent plant community (Saguaro National Park, Arizona, USA). To inform our strength of inference, we evaluated the effects of two different nutrient media, incubation temperatures, and collection seasons on the apparent structure of endophyte assemblages. We complemented our community-level focus with phylogenetic analyses of the most frequently isolated genus (Preussia) to determine how endophytes in the Sonoran Desert's under-explored mycota can contribute to current perspectives on diversity and substrate use in a relatively well-studied clade [3, 4, 12, 19].
We use our data to test four predictions: (1) Endophytes inhabiting these desert plants will demonstrate low abundance and diversity relative to their expected latitudinal position, consistent with the relatively severe abiotic environment in which they occur [8, 9, 11, 50, 83]. (2) Endophytes of these desert plants will be host-, tissue-, and substrate-use generalists, consistent with a relatively harsh environment that may select for opportunistic symbioses [39; see also 47, 48, 69, and discussion below]. (3) Endophytes will demonstrate little spatial structure over the relatively small geographic scale of our study, consistent with the general uniformity and contiguousness of the plant community and the small spatial scale of our sampling [23, 37]; however, endophyte abundance, diversity, and community composition will differ as a function of exposure, consistent with meaningful variation in microclimate [50, 87]. (4) In a focal genus (Preussia), endophytism in desert plants will represent a single evolutionary origin, and ecological traits and the capacity to grow at particular temperatures or on particular media will be structured phylogenetically.
Methods
Fresh plant material was collected in October 2011 along three 100 m transects in each of two areas of the Tucson Mountain District of Saguaro National Park, Arizona (i.e., Saguaro West: ca. 32.25°N, 111.19°W; 793 m.a.s.l.): King Canyon Trail (KC), an d near the Red Hills Visitor Center (RH; Table 1). Surveys were repeated in the same sites in February 2012, when an additional transect was added at the RH site (i.e., seven transects total; Table 1). Collection sites were chosen for accessibility and the presence of focal species in each exposure classification (see below), but otherwise represent the typical plant communities, soil types, and land use history of this protected zone [71]. At their nearest points, the two collection sites are ca. 3 km apart.
Table 1. Collection information for surveys of endophytes in leaves and stems of Sonoran Desert plants.
collection sites, transects (labeled by exposure), coordinates at the central collecting point (lat, latitude; long, longitude), mean elevation, and host species collected only in October 2011 (*), only in February 2012 (‡), or in both collection periods (no mark). Leaves were collected from L. tridentata and S. chinensis; stems were collected from all species.
| Site | Transect | Lat (°) | Long (°) | Elevation (m) | Host species |
|---|---|---|---|---|---|
| King Canyon | Flat | 32.254 | -111.152 | 996.2 ± 5.1 | Larrea tridentata |
| Parkinsonia microphylla | |||||
| Phoradendron californicum* | |||||
| Simmondsia chinensis | |||||
| S/W | 32.255 | -111.151 | 1041 ± 3.7 | Larrea tridentata | |
| Parkinsonia microphylla | |||||
| Simmondsia chinensis | |||||
| North | 32.252 | -111.151 | 1048.7 ± 9.6 | Larrea tridentata | |
| Parkinsonia microphylla | |||||
| Simmondsia chinensis | |||||
| Red Hills | Flat-1 | 32.252 | -111.186 | 783.5 ± 33.9 | Larrea tridentata |
| Parkinsonia microphylla | |||||
| Phoradendron californicum | |||||
| Simmondsia chinensis | |||||
| Flat-2 | 32.363 | -111.212 | 731.1 ± 1.8 | Larrea tridentata‡ | |
| Parkinsonia microphylla‡ | |||||
| Phoradendron californicum‡ | |||||
| S/W | 32.251 | -111.184 | 831.3 ± 1.0 | Larrea tridentata | |
| Parkinsonia microphylla | |||||
| Simmondsia chinensis | |||||
| North | 32.252 | -111.186 | 823.3 ± 17.9 | Larrea tridentata | |
| Parkinsonia microphylla | |||||
| Simmondsia chinensis |
Saguaro West encompasses ca. 9700 ha of arid mountains, bajadas, and valleys dominated by Arizona upland Sonoran Desert scrub, typical of the northeastern zone of the Sonoran Desert biome. The most common and conspicuous woody plants include creosote (Larrea tridentata, Zygophyllaceae), jojoba (Simmondsia chinensis, Simmondsiaceae), foothills palo verde (Parkinsonia microphylla, Fabaceae), and diverse Cactaceae (especially Platyopuntia spp., Cylindropuntia spp., and Carnegiea gigantea) [71]. Annual precipitation averages 260 mm, and the mean annual temperature is approximately 21°C [2]. In October, the average high in the region is 29.9 °C, the average low is 12.7° C, and average precipitat ion is 19.1 mm [2]. Rainfall in the previous three months averages ca. 135 mm [2]. In February, the average high is 20.5 °C, the average low is 4.6°C, average monthly precipitation is 21.3 mm, and rainfall in the preceding three months averages ca. 67 mm [2].
In each sampling event, we collected plant material along three transects per sampling site: one in a flat, exposed location, one on a south- or west-facing slope (highly exposed), and one on a north-facing slope (protected) (Table 1). The transect added at RH in 2012 was flat and exposed, and was ca. 1 km from the nearest transect in that sampling site.
Each transect consisted of three collection points ca. 15 m in diameter. Points were spaced ca. 30 m apart. In each point, three individual branches (each ca. 10 cm long) were collected from one individual of each focal host species (L. tridentata, S. chinensis, and Pa. microphylla). Where possible, we also collected desert mistletoe (Phoradenron californicum; Santalaceae), a locally abundant hemiparasite of fabaceous trees. A total of 115 host plant individuals in 21 collection points (18 of which were sampled twice; Table 1) were sampled and mapped using GPS. Survey details, organized by host individual, are shown in Supplementary Table 1.
Tissue processing for endophyte isolation
Branches were placed in sealable bags and transported to the laboratory for processing. All tissue was stored at 4 °C and processed within 36 hours of col lection. Leaves (L. tridentata, S. chinensis) and stems (all focal species) were first rinsed in clean water to eliminate surface debris, and then surface-sterilized by sequential immersion in 95% ethanol (10 sec), 10% Clorox bleach (0.53% NaOCl-; 2 minutes), and 70% ethanol (2 minutes) [9, 37-40, 85-87]. Leaves of Pa. microphyllum were not processed because leaves were relatively rare and, when present, comprised leaflets that were smaller than our target tissue size.
Surface-sterilized leaves and photosynthetic stems were cut into approximately 2 mm2 pieces under sterile conditions. Care was taken to ensure that tissue fragments from leaves and stems represented similar biomass and surface area. Tissue pieces were allowed to surface-dry under sterile conditions prior to placement onto nutrient media.
Ninety-six pieces of each tissue type per individual were placed individually into 1.5 mL tubes containing 2% malt extract agar (Amresco, Solon, OH, USA) or BBL cornmeal agar (Becton, Dickinson, and Co., Franklin Lakes, NJ, USA) [37-40, 85-87], for a total of 192 leaf- and 192 stem pieces per host individual per sampling event. Tubes were sealed for incubation following [85-88]. Half of the tubes were incubated at room temperature (ca. 21.5 °C) and half at 30.3 °C (average temperature of Tucson in July; [2]). Overall, we processed 22,656 tissue segments. Emergent fungi were subcultured, vouchered in sterile water, and accessioned as living samples at the Robert L. Gilbertson Mycological Herbarium at The University of Arizona (accessions MYCO-ARIZ SNP0001-0459).
Molecular analyses
Total genomic DNA was extracted from each isolate following [9]. The polymerase chain reaction (PCR) was used to amplify the nuclear ribosomal internal transcribed spacers and 5.8S gene (ITSrDNA) and an adjacent portion of the ribosomal large subunit (LSUrDNA) for a total of ca. 1000 base pairs per sequence (i.e., ITSrDNA-LSUrDNA) [87]. PCR methods followed [37] with primers ITS1F and LR3. When amplification with these primers failed, we amplified only ITSrDNA using primers ITS1F or ITS5 and ITS4 [86].
PCR products were verified by gel electrophoresis, cleaned using ExoSap-IT (Affymetrix; Santa Clara, CA, USA), and sequenced bidirectionally using the Applied Biosystems BigDye Terminator v 3.1 cycle sequencing kit and the original PCR primers on an Applied Biosystems 3730xl DNA Analyzer (Foster City, CA, USA) at the University of Arizona Genetics Core. DNA sequences were assembled in phred [24] and phrap [25] with orchestration by Mesquite v. 1.06 [57] and then manually verified and edited in Sequencher v. 5.1 (Gene Codes Corporation, Ann Arbor, MI, USA). Overall, we successfully amplified and sequenced ITSrDNA or ITSrDNA-LSUrDNA for 457 isolates, comprising our entire collection. All sequences have been deposited at GenBank (accessions XXXXXX-XXXXXX).
Operational taxonomic units corresponding to 95%, 99%, and 100% sequence similarity were delimited using Sequencher v. 5.1 following [9]. Similarity groups based on 95% ITS rDNA-LSU rDNA similarity often approximate species boundaries in the groups of taxa recovered here [86]. All analyses described below were conducted using operational taxonomic units based on 95% sequence similarity (OTU), but results were qualitatively similar using 99% and 100% similarity groups (data not shown). OTU designations were validated by comparison with phylogenetic results in several genera, of which one is shown here (Preussia, described below).
Isolation frequency
Isolation frequency was defined as the percent of tissue segments yielding an endophyte in culture. A Wilcoxon test revealed significant differences in isolation frequency as a function of tissue type, such that the data were partitioned into two data sets prior to analysis: one representing collections from leaves of L. tridentata and S. chinensis, and one representing collections from stems of L. tridentata, S. chinensis, and Pa. microphyllum. Ph. californicum was not included in statistical analyses of isolation frequency due to its rarity in our sites.
Prior to analysis, we confirmed that isolation frequencies for leaves vs. stems of individual plants were not correlated, and that the data were not structured by sampling points. We then used generalized linear models to examine variation in isolation frequency from leaves and stems as a function of ecological variables (sampling site, plant species, sampling month, and exposure) and methodological variables (nutrient medium and incubation temperature). The sampling site was defined as KC or RH. Exposure was defined by the slope and aspect of each transect (flat, exposed; south- or west-facing, highly exposed; north, protected). The final model for each data set used a Poisson distribution, a log-link function, and isolation frequencies that were back-calculated to count data, and incorporated all main effects and relevant interaction terms (Table 2). Analyses were performed in JMP v. 10.0.0 (SAS Institute, Cary, NC, USA).
Table 2. Interaction terms define the relationship of isolation frequency from each tissue type to ecological factors, but not to isolation approaches (nutrient medium, incubation temperature).
Results of generalized linear models for analyses of isolation frequency from (A) leaves and (B) photosynthetic stems of focal Sonoran Desert plants. Whole-model test for (A): chi-square = 73.6, DF = 15, P <0.0001; AICc = 295.98; whole-model test for (B): chi-square = 165.32, DF = 37, P <0.0001; AICc = 1154.60.
| A. Source | DF | Chi-square | P |
|---|---|---|---|
| Site | 1 | 1.99 e-7 | 0.9996 |
| Species | 1 | 2.31 e-8 | 0.9999 |
| Medium | 1 | 0.40 | 0.5285 |
| Temperature | 1 | 2.70 | 0.1002 |
| Sampling month | 1 | 24.82 | <.0001* |
| Exposure | 2 | 2.84 | 0.2416 |
| Site*Species | 1 | 19.02 | <.0001* |
| Site*Sampling month | 1 | 1.33 e-8 | 0.9999 |
| Site*Exposure | 2 | 4.26 | 0.1186 |
| Sampling month*Exposure | 2 | 1.08 | 0.5835 |
| Species*Sampling month | 1 | 7.44 e-7 | 0.9993 |
| Site*Species*Sampling month | 1 | 6.0 | 0.0146* |
|
| |||
| B. Source | DF | Chi-square | P |
|
| |||
| Site | 1 | 1.95 | 0.1627 |
| Species | 2 | 1.04 | 0.5933 |
| Medium | 1 | 0.09 | 0.7601 |
| Temperature | 1 | 1.75 | 0.1855 |
| Sampling month | 1 | 10.36 | 0.0013* |
| Exposure | 2 | 2.27 | 0.3210 |
| Site*Species | 2 | 10.79 | 0.0045* |
| Site*Sampling month | 1 | 4.55 | 0.0329* |
| Site*Exposure | 2 | 0.86 | 0.6489 |
| Species*Sampling month | 2 | 9.6 | 0.0072* |
| Species*Exposure | 4 | 22.45 | 0.0002* |
| Sampling month*Exposure | 2 | 9.34 | 0.0094* |
| Site*Species*Sampling month | 2 | 7.67 | 0.0216* |
| Site*Species*Exposure | 4 | 5.78 | 0.2158 |
| Species*Sampling month*Exposure | 4 | 8.96 | 0.0620 |
| Site*Sampling month*Exposure | 2 | 0.44 | 0.8019 |
| Site*Species*Sampling month*Exposure | 4 | 11.3 | 0.0234* |
Indicate P ≤ 0.05
Richness and diversity
Species richness and the completeness of sampling were evaluated using species-accumulation curves inferred with 50 randomizations of species order in EstimateS 8.0.0 [21]. Diversity was measured as Fisher's alpha, which is robust to variation in sample size [26; see also 38-40, 85-87]. Because isolation frequency was very low, diversity could not be calculated meaningfully for all samples. Therefore, data were pooled with respect to incubation temperature, nutrient medium, sampling site, exposure, and sampling month, as none of these variables described meaningful variation in the data. The resulting analysis examined the relationship of Fisher's alpha to tissue type (leaves, stems), host species (L. tridentata, S. chinensis), and the host species × tissue type interaction term (Table 3). A Shapiro-Wilk test was used prior to analysis to confirm that the distribution of Fisher's alpha values did not differ significantly from normal. The analysis did not include Pa. microphylla, for which leaves were not sampled, or Ph. californicum, for which leaves were not available.
Table 3. Endophyte diversity differs among host species but not as a function of tissue type.
Result of multiple regression examining the relationship of diversity (Fisher's alpha) to host species (Larrea tridentata and Simmondsia chinensis) and tissue type; R2 = 0.52.
| Source | DF | Sum of Squares | F Ratio | P |
|---|---|---|---|---|
| Species | 1 | 160.39 | 6.3464 | 0.0453* |
| Tissue | 1 | 5.46 | 0.2160 | 0.6585 |
| Species*Tissue | 1 | 6.73 | 0.2664 | 0.6242 |
Indicate P ≤ 0.05
Latitudinal comparisons of isolation frequency and diversity
To determine whether isolation frequency and diversity differed from values that were expected given the latitudinal position of the study region, mean values for each were compared against expected values derived from regressions presented in [9, 11]. These studies were chosen for comparisons because they span a wide latitudinal range, focus primarily on above-ground tissues of woody plants, and use the same sampling approach as the present study (i.e., similar depth of sampling and the same nutrient media, tissue processing approach, and surface-sterilization process). Analyses were conducted twice: once using data from leaves and stems, and once using leaves only (to match previously published data, which focused on endophytes from leaves [9, 11]).
Analyses of community composition
Differences in community composition were evaluated using analyses of similarity (ANOSIM) conducted in PAST v. 1.88 [33]. Because isolation frequency was low, data were pooled prior analysis with respect to incubation temperature, nutrient medium, sampling site, sampling month, exposure, and tissue type. Singletons were removed prior to analysis. The final data set was analyzed to determine whether community composition of endophytes differed as a function of host species, as no other explanatory variable was sufficiently informative either alone or in combination (data not shown). For ANOSIM, distances (defined as 1-Morisita's index of similarity) were converted to ranks and the test statistic R was calculated as the difference of mean ranks between vs. within groups. Significance was computed by 10,000 permutations of group membership. Similarities in fungal community composition were visualized using non-metric multidimensional scaling (NMDS), an ordination method that uses rank-order information in a dissimilarity matrix [27].
These analyses were complemented by comparisons of mean pairwise similarity values for endophyte communities in conspecific vs. heterospecific hosts. Similarity values (Jaccard's index, based on presence/absence of nonsingleton OTU; Morisita-Horn index, based on abundance of nonsingleton OTU) were calculated in EstimateS [21] using all possible pairwise comparisons for host individuals from which ≥4 isolates were obtained, and compared between conspecific and heterospecific hosts using t-tests of logit-transformed data. Mean pairwise similarity was compared against expected values for temperate ecosystems based on [9, 11] to determine if host affiliations were consistent with expected values given the latitudinal position of the study region.
Phylogenetic analyses
Examination of cultures and BLASTn results indicated that the most commonly isolated genus in our sample was Preussia (Sporormiaceae, Pleosporales, Dothideomycetes, Ascomycota; at least 121 isolates representing multiple OTU). Preussia is known primarily from dung and leaf litter [3, 12, 19] but also contains endophytic species [4, 39, 40, 69]. We inferred the phylogenetic relationships of endophytic Preussia obtained here in the context of currently recognized species for which sequence data are available [see 3, 4, 12, 19] to validate OTU designations, to examine the contribution of these desert strains to the currently recognized structure of the genus, and to determine whether endophytes in this genus reflect a single evolutionary origin and have phylogenetically structured traits relevant to isolation in culture (i.e., preferred growth medium and incubation temperature) and their ecological distributions.
Sequence data used in recent phylogenetic studies and comprising the majority of known species of Preussia were obtained from GenBank (Supplementary Table 2) and aligned with all isolates from the present study that were identified as Preussia by BLASTn (Supplementary Table 3) and preliminary phylogenetic analyses [20, 39, 40]. The resulting data set was trimmed to consistent start- and end points, and for maximum taxonomic richness it was confined only to ITSrDNA (final length, 560 base pairs). Redundant sequences were removed and the resulting data set of 110 terminals, including the outgroup (Pleospora herbarum, as specified by [4]) was aligned using MUSCLE [94] and then manually edited in MacClade v. 4.08a [56].
The resulting alignment was analyzed using maximum likelihood analyses in GARLI [95] and Bayesian analyses in MrBayes v. 3.1.2 [42] with the GTR+I+G model of evolution implemented in each case based on BIC results from jModelTest [70]. The Bayesian analysis consisted of 3 million generations, sampling every 1000th tree, with four chains and a random starting tree. Completion was assessed by examining-ln li values and the standard deviation of split frequencies. A majority rule consensus tree was inferred from 500 trees from the posterior, with support defined by Bayesian posterior probabilities. Complementary support values were provided by 100 maximum likelihood bootstrap replicates conducted as above in GARLI.
Results
Surveys of common Sonoran Desert plants in two areas of Saguaro National Park (Arizona, USA) yielded 457 isolates of fungal endophytes from 22,656 tissue segments representing photosynthetic tissues of Larrea tridentata, Parkinsonia microphylla, Phoradendron californicum, and Simmondsia chinensis (isolation frequency = 2.0%).
Isolation frequency differed significantly as a function of tissue type. Endophytes were isolated in culture ca. four times more frequently from stem tissue than from leaf tissue overall (all species), and 3.7 times more frequently in species from which both leaves and stems were considered (L. tridentata and S. chinensis; chi-square = 65.07, DF = 1, P < 0.0001).
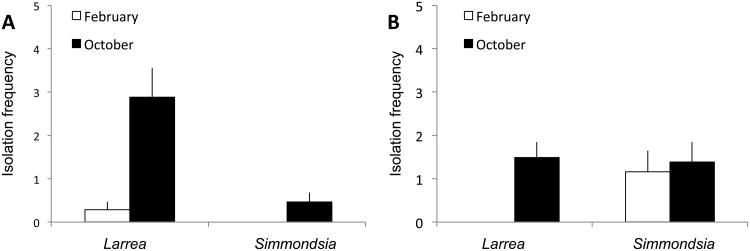
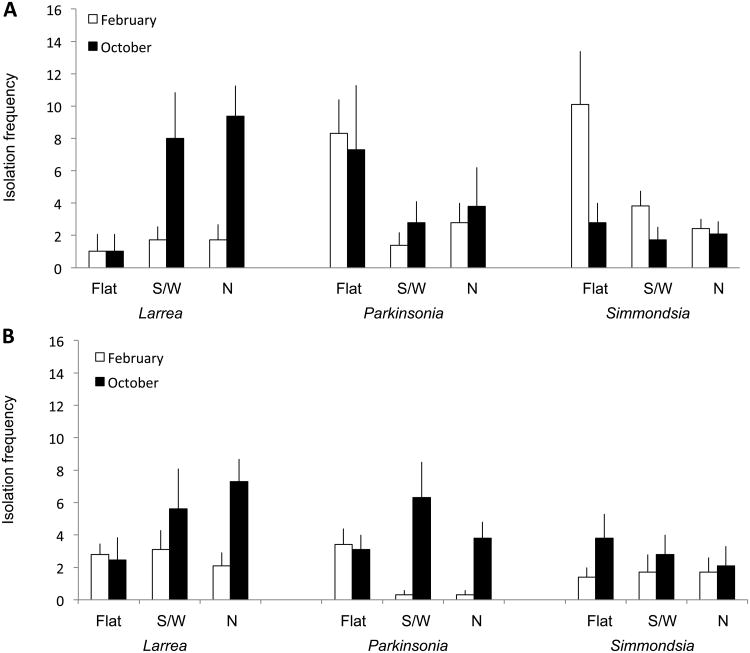
Isolation frequency did not differ as a function of incubation temperature or nutrient medium for either tissue type (Table 2). However, isolation frequency for leaves differed as a function of a three-way interaction (sampling site × species × sampling month; Table 2, Fig. 1). Isolation frequency for stems varied as a function of a four-way interaction (sampling site × species × sampling month × exposure; Table 2, Fig. 2).
Fig. 1.
Isolation frequency for endophytes from leaves of Sonoran Desert plants reflects a site × species × sampling month interaction (see Table 2). Data indicate the percent of tissue segments from leaves of Larrea tridentata and Simmondsia chinensis that yielded endophytes in culture as a function of sampling site (A, King Canyon; B, Red Hills Visitor Center) and sampling month (October vs. February).
Fig. 2.
Isolation frequency for endophytes from photosynthetic stems of Sonoran Desert plants reflects a site × species × sampling month × exposure interaction (see Table 2). Data indicate the percent of tissue segments from stems of Larrea tridentata, Parkinsonia microphylla, and Simmondsia chinensis that yielded endophytes in culture, as a function of sampling site (A, King Canyon; B, Red Hills), sampling month (October vs. February), and exposure (flat, exposed; south- or west-facing, highly exposed; or north-facing, protected).
Despite low abundance, cultivable endophytes were highly diverse. Overall, 89 OTU were obtained from 457 sequenced isolates (Fisher's alpha = 31.8; OTU based on 95% sequence similarity). Of these, 46 (51.6%) occurred only once. Overall, 201 distinct genotypes were obtained (i.e., based on 100% sequence similarity).
The species accumulation curve was non-asymptotic, but comparison of observed species richness and the bootstrap estimate suggests that ca. 83% of available, cultivable OTU were isolated during our surveys (Fig. 3), thus providing a robust basis for the analyses described below.
Fig. 3.
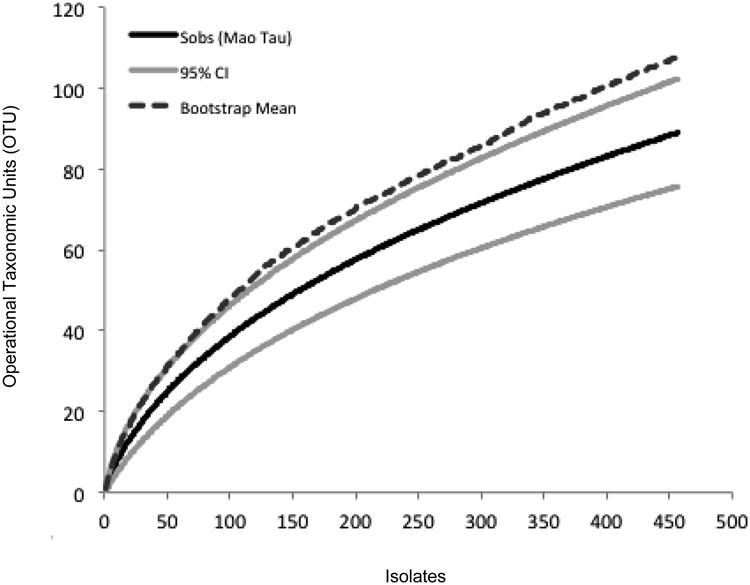
Species accumulation curve reveals high richness of cultivable fungal endophytes in leaves and stems of four common species of non-succulent woody plants in two collection sites in the Sonoran Desert (Tucson Mountain District, Saguaro National Park). Solid black line: number of observed OTU (Sobs, based on 95% sequence similarity); grey lines: upper and lower bounds of the 95% confidence interval around observed richness; dashed line: bootstrap estimate of total species richness.
Endophyte diversity did not differ as a function of tissue type in the plants for which both leaves and stems were sampled, and there was no evidence for a tissue type × species interaction (Table 3). However, diversity did differ significantly among host species, ranging from a mean Fisher's alpha of 7.9 ± 1.9 (S. chinensis) to 15.7 ± 4.8 (L. tridentata) (Table 3, Table 4).
Table 4. Desert endophytes are rare in culture but highly diverse.
Isolation frequency was significantly lower than expected given the latitude of the study site, whereas diversity from each host species did not differ from the expected value given latitude. Expected values are means for collections at 30-36°N as presented in refs. 9 and 11 (mean ± S D, and 95% confidence interval). The final column indicates whether the value observed here was significantly higher or lower than the expected value (Z-test).
| Measurement | Relevant data: mean ± SD | Expected value (95% CI) | Mean significantly lower/higher than expected (P-value) |
|---|---|---|---|
| Isolation frequency* | Leaves, 0.9 ± 0.4 | 40.2 ± 29.4 (8.9-59.9) | Lower than expected; (P<0.0001)‡ |
| Stems, 3.5 ± 1.2 | |||
| Diversity (Fisher's alpha)** | Larrea, 15.7 ± 4.8 | 11.6 ± 4.0 (8.6-14.8) | No difference from expected; (P>0.05)‡ |
| Parkinsonia, 11.4 ± 3.5 | |||
| Simmondsia, 7.9 ± 1.9 |
Isolation frequency differed significantly as a function of tissue type; see text and Table 2.
Diversity differed among host species but not as a function of other explanatory variables; see Table 2.
Result was consistent when based on data from leaves only, and when based on data from leaves and stems.
As predicted, mean isolation frequencies from leaves were significantly lower than expected given the latitude of the study region (Table 4). However, mean diversity values from leaves were consistent with expected values (Table 4). These patterns remained consistent when data from stems were included (Table 4).
ANOSIM revealed that the composition of endophyte communities differed markedly among host species (Fig. 4). Community similarity was significantly higher among samples from conspecific hosts vs. heterospecific hosts (Fig. 4). In contrast to our prediction, mean similarities were consistent with expected values based on the latitudinal position of the study region (Fig. 4). However, the majority of OTU that were isolated more than once occurred in more than one host species (Supplementary Table 4).
Fig. 4.
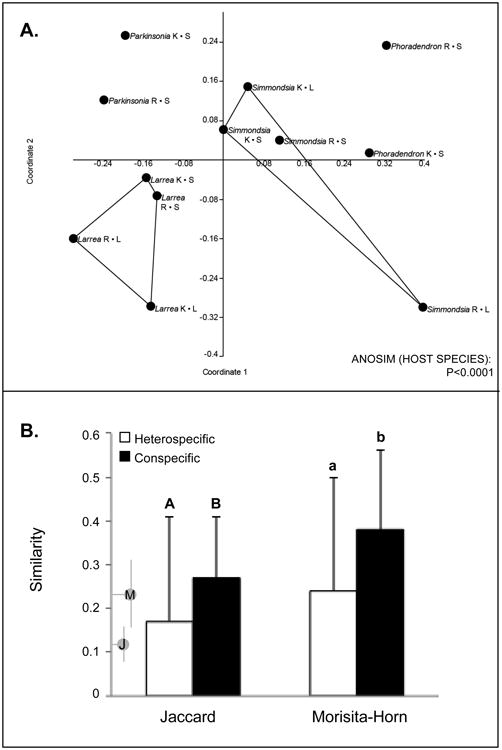
Community-level analyses reveal significant structuring of endophyte communities by host species. (A) Non-metric multidimensional scaling of endophyte communities and ANOSIM results based on the Morisita index, calculated using nonsingleton OTU. (B) mean pairwise similarity values for endophyte communities in conspecific vs. heterospecific hosts, calculated using Jaccard's index and the Morisita-Horn index (nonsingletons only, and only including host individuals for which ≥4 endophytes were obtained in culture). Annotations on the y-axis indicate mean values (dots) and ranges (bars) expected for temperate comparisons using Jaccard's index (J) and the Morisita-Horn index (M) (values obtained from [9]).
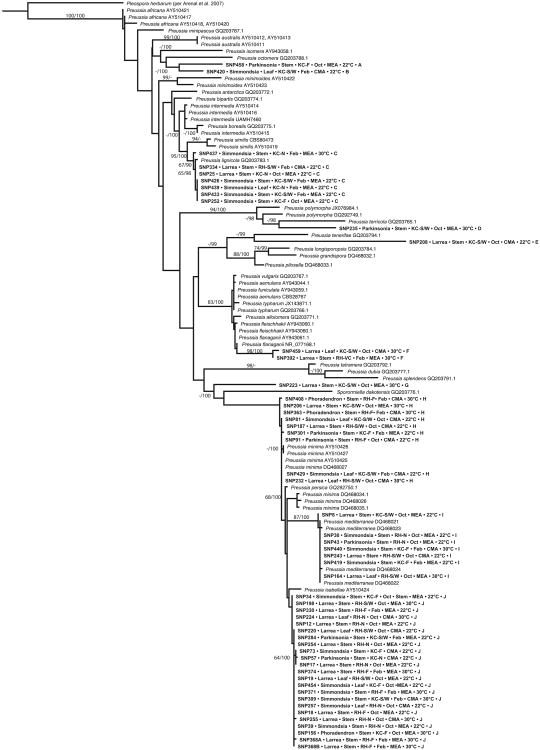
Concomitant with their high species richness, endophytes were phylogenetically diverse. Among 89 OTU we recovered representatives of four classes of Pezizomycotina (Ascomycota) in culture (Dothideomycetes, Eurotiomycetes, Pezizomycetes, and Sordariomycetes) (Supplementary Table 3). We also isolated several strains with affinity for the Mucoromycotina (incertae sedis). Dothideomycetes were especially common, with particular prevalence by Preussia in terms of abundance and species richness (Supplementary Table 3; Fig. 5).
Fig. 5.
Phylogenetic analysis of Preussia endophytes obtained from Sonoran Desert plants reveals high phylogenetic diversity, congruence of clades with OTU, and a lack of structure with regard to ecological and methodological factors. Terminal taxa in bold were isolated in this study and are annotated to indicate isolate number, host, tissue of origin, site and transect, sampling month, isolation medium and temperature, and OTU group, here listed such that shared letters indicate membership in the same OTU. Tree depicts the results of maximum likelihood (ML) analysis of ITSrDNA data; support values are ML bootstrap values (before slash; values ≥60% are shown) and Bayesian posterior probabilities (after slash; values ≥ 90% are shown).
Phylogenetic analyses
Preussia spp. were the top BLAST matches for ca. 25 isolates overall, but 121 isolates could be placed in the genus using phylogenetic methods (Fig. 5; Supplementary Table 3). Phylogenetic analyses revealed a high phylogenetic richness of Sonoran Desert endophytes within Preussia (Fig. 5). Several clades were affiliated closely with known species (e.g., Pr. isabellae, Pr. lignicola, Pr. mediterranea, and Pr. minima), thus expanding the known geographic range and substrate use of these clades. At least six strains or clades were distinct from previously sequenced species (Fig. 5). Ten clades of Preussia obtained here corresponded directly to OTU (Fig. 5), corroborating the robustness of our OTU designations for ecological analyses.
Endophytes associated with the plants surveyed here did not form a single, monophyletic group, but instead were interspersed with known taxa from other regions and different substrates (e.g., dung- and leaf-litter affiliates; Supplementary Table 2). No phylogenetic pattern was evident with regard to the medium on which a strain was isolated, or the capacity to be isolated at the temperatures used here (but see the Pr. lignicola clade, Fig. 5). Consistent with community-level analyses, we observed no phylogenetic structure with regard to the tissue, sampling site, exposure, or sampling month from which isolates were obtained. Although community analyses revealed a strong effect of host species, endophytes in the genus Preussia showed little structure in terms of host affiliation (but see the Pr. lignicola clade, Fig. 5).
Discussion
Fungal symbionts of plants, including endophytes in above-ground tissues, are increasingly recognized for their important roles in shaping plant physiology and ecology, and for their applications in human health and sustainability [13, 22, 30, 32, 35, 65, 75, 80]. The goal of our study was to examine the abundance, diversity, local distributions, and host affiliations of endophytes in leaves and stems of representative woody plants in the Sonoran Desert, North America's hottest and most species-rich desert biome [71]. The resulting data set complements a growing body of work on root- and shoot-associated endophytes of grasses and other common plants in deserts and grasslands [1, 15-18, 31, 34, 47, 48, 50, 52-55, 59, 67-69, 82, 83, 92], providing insights into the factors shaping endophyte ecology and plant-fungal associations in diverse arid- and semi-arid lands.
Low isolation frequency
Isolation frequency of horizontally transmitted endophytes in above-ground tissues is often negatively correlated with UV radiation and aridity [8, 9, 50, 87]. Therefore, we predicted that endophytes would be isolated rarely, and that isolation frequencies would be low relative to expected values given the latitudinal position of our study region (prediction 1).
We found that endophytes associated with stems and leaves were isolated roughly eleven- to forty-fold less frequently than expected given the latitude of our study site (Table 4). On average, endophytes were isolated in culture from only ca. 1% of leaf segments and 3.5% of stem segments (Table 4). We do not attribute this low isolation frequency to a detrimental artifact of our surface-sterilization approach; previous studies using the same method have recorded much higher isolation frequencies in other biomes, including local mountains with more mesic conditions [9, 11, 35, 37, 39, 85-88]. Instead, we anticipate that it reflects a relatively low volume of endophyte inoculum, as evidenced by low rates of sporefall in the Sonoran Desert compared to more mesic biomes (Arnold, unpubl. data; see also [5]). Moreover, the conditions under which endophytic symbioses establish readily in experimental and natural settings, including high humidity, moderate temperatures, high plant density, and low solar- or UV radiation [8, 10, 50], are not common in the study region.
Many factors may influence isolation frequency from particular tissues. In above-ground tissues of woody plants, endophyte isolation frequency is often positively correlated with tissue age [10]. Here, we observed ca. four-fold higher isolation frequency from stems vs. leaves (Table 4). We predict that this difference reflects tissue age rather than effects of pervasive differences in tissue structure or chemistry, as diversity and community composition did not differ between these two tissue types (see below; see also Table 3 and Fig. 5). We could not reliably estimate the ages of stem- and leaf tissues in these wild plants, but stems in these species are longer-lived than leaves [84] and, based on relative positions, are older than the leaves they subtend.
Sensitivity of isolation frequency to environmental factors was revealed in the present study through interaction terms for both tissue types, suggesting complex relationships to local conditions and species traits. Once effects of study site and host species were taken into account, isolation frequency was generally higher for leaves in October, following a three-month period characterized by relatively high humidity and rainfall driven by the North American Monsoon, than in February, for which the preceding months are drier and cooler (Fig. 1). In stems, a more complex pattern reflected variation shaped by the interaction of study site, host species, sampling month, and exposure (Fig. 2). For stem samples, isolation frequency values in February rarely exceeded those in October, but effects of exposure varied unpredictably among species and between study sites. Such heterogeneity merits further study.
Importantly, our inferences are limited because we focused only on endophytes that could be isolated in culture, and specifically, on those that could be cultured on malt extract and cornmeal agars. A growing body of research indicates that like most microbiomes, those of plants contain many species that have not yet been cultured and may only be observed using culture-free approaches [7, 14, 38, 44-46, 54, 88, 93]. Such approaches may be especially important in harsh environments: in such cases, obligate symbioses may be favored and are less likely to be characterized readily by culturing (but see [34, 47, 69, 88] and discussion below). Thus culture-free methods are warranted in future work. At present, our study contributes publically available cultures that can be used to diagnose new lineages or population structure based on multi-locus data sets [20, 29, 64], evaluate roles of endophytes in plant health or metabolite production [13, 22, 30, 32, 35, 65, 75, 80], and generate hypotheses for culture-independent, next-generation studies.
High species richness and phylogenetic diversity
Much like isolation frequency, the diversity of cultivable, horizontally transmitted endophytes in above-ground tissues is often negatively associated with factors such as UV radiation and aridity [8, 9, 50, 87]. We predicted that endophytes inhabiting photosynthetic tissues of these desert plants would not be highly diverse, and would exhibit lower diversity than expected given the latitude of the study region (see prediction 1). Despite low isolation frequency, however, endophytes were species-rich and phylogenetically diverse both at a community level and within the most commonly isolated genus (Table 4, Fig. 5). Although next-generation methods are likely to reveal a greater number of species [7, 38, 54, 67, 88], the present study revealed at least 89 putative species among 457 isolates – a high richness given the aridity of the site and our inclusion of only four plant species and two proximate study sites. These endophytes represented more than 200 distinct ITSrDNA or ITSrDNA-LSUrDNA genotypes, suggesting that population-level studies of these fungal communities may be especially enlightening (e.g., [64]) and may reveal structure as a function of environmental or cultural characteristics that could not be resolved with OTU based on ribosomal sequences alone.
In turn, the particular prevalence of Dothideomycetes, Eurotiomycetes, and Sordariomycetes observed here (Supplementary Table 3) resembles that observed in other arid-land studies, including those of roots and non-woody plants [34, 39, 47, 52, 67, 69] and above-ground tissues of cacti [17, 83]. In general we hesitate to assign taxonomic information in the absence of phylogenetic or extensive morphological analysis, but BLASTn results suggest that in addition to Preussia, common genera likely included Phoma, Aureobasidium, Botryosphaeria, Penicillium, Aspergillus, Chaetomium (Supplementary Table 3). These taxa often are well-represented among endophytes in foliage and photosynthetic stems of woody angiosperms in many environments [9, 11, 87], but many of the putative species observed here are distinct from the majority reported in previous work. Overall, sequences obtained for 48 of 89 OTU (53.9%) were ≤97% similar to previously sequenced strains in GenBank. When we excluded OTU for which the top one or two BLAST matches were to endophytes from our previous work (e.g., [9, 11, 39, 85-88]), 22 of 35 remaining OTU (62.9%) were ≤97% similar to previously sequenced strains. This is comparable to the novelty described via culture-free methods by Porras-Alfaro et al. [67] in their examination of root-associated fungi in a desert grass.
Endophyte communities can be shaped by plant genotype [14] and by extension might be expected to exhibit diversity that is positively correlated with plant community richness [37, 50, 77, 87]. Here, high species richness and concomitantly high diversity may reflect the botanical richness and phylogenetic diversity of the Sonoran Desert bioregion [71] and host affinity of endophytes (as demonstrated by our community analyses; see below). Although above-ground tissues might be expected to harbor a lower species richness than roots in desert plants, we found up to 54 OTU associated with leaves and stems of a single species (L. tridentata, from which 171 isolates were obtained; Supplementary Table 4), comparing favorably to previous work on roots of desert plants [67].
Generalism in host-, tissue, and substrate use (prediction 2)
Symbioses often allow symbiotic partners to mitigate or overcome stress from biotic and abiotic challenges [10, 30, 41, 43, 60, 61, 73-75]. Given the expected sensitivity of many fungi to high temperatures, desiccation, and other stressors associated with this desert environment, the benefits of symbiosis over free-living status could select for opportunism and generalism in colonizing available host tissue [39; see also 47, 48, 69]. Alternatively, severe environmental conditions could select for host-specific associations, especially if such associations confer benefits on both partners -- or provide one partner with sufficient benefit, and the other with only a limited cost of association. It is also possible that host affinity could occur without strong benefits, and instead could reflect functional differences in phenology, chemistry, or other traits that differ among fungi or host plant taxa [8, 11, 45, 55, 65, 78].
We found strong evidence that endophyte communities differed among host species (Fig. 4). However, this result was not specific to these desert plants: mean community similarities within and between species resemble those in temperate areas where conditions are more mesic and plant density and diversity are often higher (Fig. 4) [9, 11]. Thus, we do not see a stronger or weaker degree of host affinity than would be expected given the latitudinal position of our study region.
The host affinity observed here contrasts with that reported in previous work on root endophytes in grasslands [47, 48], but are consistent with studies in above-ground woody plants in arid systems [50]. Overall, nonsingleton OTU were typically found in more than one host species (Supplementary Table 4; mean host range, 2.5 host taxa) and the two most common OTU were found in all hosts (OTU 2 and 7, corresponding to Preussia and Phoma, respectively; Supplementary Tables 3, 4). Thus the community structure we observed reflects assemblages of fungi that differ among host species, rather than strict-sense host specificity of particular strains. Importantly, our analyses considered only nonsingleton OTU; by definition, singleton OTU, which were represented by only one isolate, were found in only one host species. The host affinity of these very rare OTU remains to be determined and would be informed by next-generation studies, in which they may be observed more frequently.
Despite evidence for host affiliations, our data at both the community level and in analyses of Preussia were consistent with tissue- and substrate-use generalism: no differences were observed in endophyte diversity or composition in leaves vs. stems, nor as a function of incubation temperature or growth medium. Previous work on root endophytes in a desert grass revealed differences in diversity as a function of tissue type [34]: in that study, endophyte diversity in crowns differed significantly from that in leaves. In the present study, the lack of differences in diversity and species composition between stems and leaves may reflect their functional similarity: stems considered here were photosynthetic, such that they likely encompass a similar degree of biochemical dynamism compared to leaves [6].
In turn, cultivation conditions generally are expected to influence inferences regarding endophyte community structure [6], but the different cultivation conditions used here did not lead to the isolation of different fungal taxa. It is possible that a temperature difference of ca. 9 °C was insufficient to en courage growth by different taxa in vitro, concomitant with our observation of consistent diversity and community structure across seasons and despite differences in exposure, which should be correlated with marked differences in microclimate [71]. Similarly, it is possible that malt extract and cornmeal agars are not sufficiently different to encourage the selective isolation of distinctive strains. Major differences in nutrient composition, as achieved by inclusion of plant extracts in media [8, 50], could be informative in future studies. Similarly, culture-free methods could be used to determine whether only a highly generalized subset of a larger fungal community is considered when studies rely on culture-based approaches.
More generally, substrate- and temperature generalism may speak to flexibility in nutrient use and growth conditions in nature, which may be a useful strategy for horizontally transmitted endophytes given low plant density and high temperatures, UV radiation, and aridity [9, 11, 39, 50, 87]. We hypothesize that many fungi in this study have the capacity to grow on diverse substrates, but are also adept at overcoming the defenses of (or otherwise utilizing) particular host taxa, thus yielding the community-level host structure we observed here. Whether such flexibility and selectivity may be intrinsic or reflect other factors remains to be evaluated. Notably, endohyphal bacteria are common in endophytes of Sonoran Desert plants [72] and can alter the nutrient use, growth optima, and thermotolerance of their fungal hosts [40, 41, 49, 66]. We predict that ecological associations of these desert fungi may be driven in part by cryptic associations with other microbes.
Spatial patterns (prediction 3)
Given the contiguous nature of the plant community in our study region and the geographic proximity of our sampling sites, we predicted that endophytes would demonstrate little spatial structure. This prediction was upheld in terms of diversity and composition, but we found differences in isolation frequency as a function of interaction terms (Table 2, Fig. 1, Fig. 2). A lack of meaningful variation in diversity and composition across our study area was corroborated by phylogenetic analyses of Preussia, which reveal no phylogenetic structure with regard to sampling site (Fig. 5).
These results contrast markedly with one of the few studies published to date that specifically references the spatial scale of turnover in local endophyte assemblages [37]. In that study, endophyte communities in tropical forest grasses decreased markedly in similarity over distances as short as 1 km. In contrast, [23] found that spatial position and community similarity were not strongly related in endophytes tropical ferns. Overall our data suggest that inter-site distances of >3 km may be needed to encounter distinct communities of endophytic fungi in above-ground tissues of these desert plants. For applied work that relies on efficient isolation of distinctive endophyte communities, our data further suggest focusing on stems rather than leaves, given the disparity we observed in isolation frequency; including multiple host species; and designing studies that encompass single survey events across large geographic areas, rather than repeated sampling across seasons or among multiple microsites within particular areas.
Evolutionary perspectives (prediction 4)
Given the distinctive features of the Sonoran Desert, which includes a high proportion of endemic plant and animal species [71, 84], we anticipated that desert-plant endophytes in a focal genus would represent a single evolutionary origin followed by in situ diversification. However, phylogenetic analyses of Preussia spp. revealed multiple clades of endophytes from these desert plants (Fig. 5). These clades are interspersed with dung-affiliated and leaf-litter species from diverse geographic regions, consistent with multiple origins of desert-plant endophytism across the genus. The topology expands our current understanding of both the scale of richness within Preussia, and the substrate use and host range of its major subgeneric lineages (Fig. 5, Supplementary Table 2).
Concordant with that phylogenetic diversity, we found no evidence for phylogenetic structure with regard to a preferred nutrient medium or incubation temperature (Fig. 5). Previous studies have revealed the ecological and evolutionary dynamism of various Pezizomycotina with regard to the relationship of endophytic and saprotrophic lineages [11, 79, 85, 86], and growing evidence suggests that saprotrophic fungi may show more flexibility with regard to host or substrate use than endophytes [63, 85]. It is possible that endophytic clades nested within a lineage comprised mostly of saprotrophic species will demonstrate greater generalism than those with endophytic, pathogenic, or other symbiotic ancestors [11]. This hypothesis remains to be tested, with systematic surveys of endophytes from diverse lineages of plants and biomes providing the necessary taxon sampling and ecological insights for doing so. Endophytes inhabiting foliage and stems of non-succulent woody plants in the Sonoran Desert, shown here to be rare in culture but highly diverse and distinctive, thus shed light not only on a previously unknown aspect of biodiversity and species interactions in arid lands, but on the ecology and evolution of the Ascomycota, the most species-rich fungal phylum.
Supplementary Material
Supplementary Table 1. Collection details, organized by host.
Supplementary Table 2. GenBank accession numbers and ecological details for known Preussia spp. used in phylogenetic analysis.
Supplementary Table 3. Collection information, OTU designations, and BLAST matches for endophytes.
Supplementary Table 4. Summary of isolation data as a function of host species.
Acknowledgments
We thank the Western National Parks Association (MMND, SHF, NCM, AEA), the National Geographic Society (MMND, SHF, NCM, AEA), the National Institutes of Health (R01-CA90265, AEA), the National Science Foundation (AEA), Science Foundation Arizona (MHW), and the College of Agriculture and Life Sciences for supporting this research. For logistical support we thank the staff of Saguaro National Park, especially Don Swann, Anna Iwaki, and Natasha Kline. We are particularly grateful to > 400 citizen scientists who interacted with our research group at the National Geographic BioBlitz in Saguaro National Park, during which the first sampling was conducted (October 2011); and to ca.120 students at Tucson High Magnet School, who contributed greatly to all aspects of the February 2012 sampling. For help in working with students from Tucson High and contributing to our group's BioBlitz outreach and research activities, we thank Brett Baxter, Lauren Dominick, Chan Jung, Adrian Ramirez, and Robert (Ethan) Posey. Support for our outreach and research activities with students and citizen scientists is gratefully acknowledged: College of Agriculture and Life Sciences at the University of Arizona, the Robert L. Gilbertson Mycological Herbarium, and Tucson High Magnet School.
References
- 1.Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Applied and Environmental Microbiology. 2012;78:7527–7537. doi: 10.1128/AEM.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. Western Regional Climate Center; 2013. http://www.wrcc.dri.edu/ [Google Scholar]
- 3.Arenal F, Platas G, Peláez F. Two new Preussia species defined based on morphological and molecular evidence. Fungal Diversity. 2005;20:1–15. [Google Scholar]
- 4.Arenal F, Platas G, Peláez F. A new endophytic species of Preussia (Sporormiaceae) inferred from morphological observations and molecular phylogenetic analysis. Fungal Diversity. 2007;25:1–17. [Google Scholar]
- 5.Arnold AE. Dissertation. The University of Arizona; 2002. Neotropical fungal endophytes: diversity and ecological roles. [Google Scholar]
- 6.Arnold AE. Understanding the diversity of foliar fungal endophytes: progress, challenges, and frontiers. Fungal Biology Reviews. 2007;21:51–66. [Google Scholar]
- 7.Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 2007;99:185–206. doi: 10.3852/mycologia.99.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: Ecological pattern and process in Theobroma cacao (Malvaceae) Mycologia. 2003;95:388–398. [PubMed] [Google Scholar]
- 9.Arnold AE, Lutzoni FL. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 10.Arnold AE, Mejía L, Kyllo D, Rojas D, Maynard Z, Herre AE. Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences USA. 2003;100:15649–15654. doi: 10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold AE, Miadlikowska J, Higgins KL, Sarvate SD, Gugger P, Way A, Hofstetter V, Kauff F, Lutzoni F. A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Systematic Biology. 2009;58:283–297. doi: 10.1093/sysbio/syp001. [DOI] [PubMed] [Google Scholar]
- 12.Asgari B, Zare R. Two new species of Preussia from Iran. Nova Hedwigia. 2010;90:3–4. [Google Scholar]
- 13.Bae H, Sicher RC, Kim MS, Kim SH, Strem MD, Melnick RL, Bailey BA. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. Journal of Experimental Botany. 2009;60:3279–3295. doi: 10.1093/jxb/erp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bálint M, Tiffin P, Hallström B, O'Hara RB, Olson MS, Frankhauser JD, Piepenbring M, Schmitt I. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera) PLoS ONE. 2013;8:e53987. doi: 10.1371/journal.pone.0053987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrow JR. Atypical morphology of dark septate fungal root endophytes of Bouteloua in arid southwestern USA rangelands. Mycorrhiza. 2003;13:239–247. doi: 10.1007/s00572-003-0222-0. [DOI] [PubMed] [Google Scholar]
- 16.Barrow JR, Osuna P. Phosphorus solubilization and uptake by dark septate fungi in fourwing saltbush, Atriplex canescens (Pursh) Nutt. Journal of Arid Environments. 2002;51:449–459. [Google Scholar]
- 17.Bezerra JDP, Santos MGS, Svedese VM, Lima DMM, Fernandes MJS, Paiva LM, Souza-Motta CM. Richness of endophytic fungi isolated from Opuntia ficus-indica Mill. (Cactaceae) and preliminary screening for enzyme production. World Journal of Microbiology and Biotechnology. 2012;28:1989–1995. doi: 10.1007/s11274-011-1001-2. [DOI] [PubMed] [Google Scholar]
- 18.Bills RJ, Stutz JC. AMF associated with indigenous and non-indigenous plants at urban and desert sites in Arizona. In: Azcón-Aguilar C, et al., editors. Mycorrhizas-Functional Processes and Ecological Impact. Springer; Berlin Heidelberg: 2009. pp. 207–220. [Google Scholar]
- 19.Cain RF. Studies of coprophilous ascomycetes VII. Preussia. Canadian Journal of Botany. 1961;39:1633–1666. [Google Scholar]
- 20.Chen KH, Miadlikowska J, Molnar K, Arnold AE, U'Ren JM, Gaya E, Gueidan C, Lutzoni F. Phylogenetic analyses of eurotiomycetous endophytes reveal their close affinities to Chaetothyriales, Eurotiales, and a new order – Phaeomoniellales. Molecular Phylogenetics and Evolution. doi: 10.1016/j.ympev.2015.01.008. in press. [DOI] [PubMed] [Google Scholar]
- 21.Colwell RK. EstimateS, Version 8.2: Statistical Estimation of Species Richness and Shared Species from Samples. 2011 http://viceroy.eeb.uconn.edu/Colwell/EstimateS.
- 22.Davis EC, Shaw AJ. Biogeographic and phylogenetic patterns in diversity of liverwort-associated endophytes. American Journal of Botany. 2008;95:914–924. doi: 10.3732/ajb.2006463. [DOI] [PubMed] [Google Scholar]
- 23.Del Olmo Ruiz M, Arnold AE. Interannual variation and host affiliations of endophytic fungi associated with ferns at La Selva, Costa Rica. Mycologia. 2014;106:8–21. doi: 10.3852/13-098. [DOI] [PubMed] [Google Scholar]
- 24.Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- 25.Ewing B, Hiller L, Wendl M, Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 26.Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. Journal of Animal Ecology. 1943;12:42–58. [Google Scholar]
- 27.Gauch HG. Multivariate analysis in community structure. Cambridge University Press; UK: 1982. [Google Scholar]
- 28.Gazis R, Chaverri P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecology. 2009;3:240–254. [Google Scholar]
- 29.Gazis R, Miadlikowska J, Lutzoni F, Arnold AE, Chaverri P. Culture-based study of endophytes associated with rubber trees in Peru reveals a new class of Pezizomycotina: Xylonomycetes. Molecular Phylogenetics and Evolution. 2012;65:294–304. doi: 10.1016/j.ympev.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 30.Giauque H, Hawkes CV. Climate affects symbiotic fungal endophyte diversity and performance. American Journal of Botany. 2013;100:1435–1444. doi: 10.3732/ajb.1200568. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert SF, McDonald E, Boyle N, Buttino N, Gyi L, Mai M, Prakash N, Robinson J. Symbiosis as a source of selectable epigenetic variation: taking the heat for the big guy. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:671–678. doi: 10.1098/rstb.2009.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunatilaka AAL. Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. Journal of Natural Products. 2006;69:509–526. doi: 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
- 34.Herrera J, Khidir HH, Eudy DM, Porras-Alfaro A, Natvig DO, Sinsabaugh RL. Shifting fungal endophyte communities colonize Bouteloua gracilis: effect of host tissue and geographical distribution. Mycologia. 2010;102:1012–1026. doi: 10.3852/09-264. [DOI] [PubMed] [Google Scholar]
- 35.Higginbotham SJ, Arnold AE, Ibañez A, Spadafora C, Coley PD, Kursar TA. Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants. PLoS One. 2013;8:e73192. doi: 10.1371/journal.pone.0073192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higginbotham SJ, Wong WR, Linington RG, Spadafora C, Iturrado L, Arnold AE. Sloth fur as a novel source of fungi with potent anti-parasitic, anti-cancer and anti-bacterial activity. PLoS One. 2014;9:e84549. doi: 10.1371/journal.pone.0084549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins KL, Arnold AE, Coley PD, Kursar TK. Communities of fungal endophytes in tropical forest grasses: highly diverse host- and habitat generalists characterized by strong spatial structure. Fungal Ecology. 2014;8:1–11. [Google Scholar]
- 38.Higgins KL, Coley PD, Kursar TA, Arnold AE. Culturing and direct PCR suggest prevalent host-generalism among fungal endophytes of tropical grasses. Mycologia. 2011;103:247–260. doi: 10.3852/09-158. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman M, Arnold AE. Geography and host identity interact to shape communities of endophytic fungi in cupressaceous trees. Mycological Research. 2008;112:331–344. doi: 10.1016/j.mycres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Hoffman MT, Arnold AE. Diverse bacteria inhabit living hyphae of phylogenetically diverse foliar endophytes. Applied and Environmental Microbiology. 2010;76:4063–4075. doi: 10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman MT, Gunatilaka MK, Wijeratne EMK, Gunatilaka AAL, Arnold AE. Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte. PLoS ONE. 2013;8:e73132. doi: 10.1371/journal.pone.0073132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 43.Janson EM, Peeden ER, Stireman JO, Abbot P. Symbiont-mediated phenotypic variation without co-evolution in an insect-fungus association. Journal of Evolutionary Biology. 2010;23:2212–2228. doi: 10.1111/j.1420-9101.2010.02082.x. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan D, Maymon M, Agapakis CM, Lee A, Wang A, Prigge BA, Volkogon M, Hirsch AM. A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev Desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. American Journal of Botany. 2013;100:1713–1725. doi: 10.3732/ajb.1200615. [DOI] [PubMed] [Google Scholar]
- 45.Kembel SW, O'Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proceedings of the National Academy of Sciences. 2014;38:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kemler M, Gamas J, Wingfield MJ, Gryzenhout M, Pillay KA, Slippers B. Ion Torrent PGM as a tool for fungal community analysis: a case study of endophytes in Eucalyptus grandis reveals high taxonomic diversity. PLoS One. 2013;8:e81718. doi: 10.1371/journal.pone.0081718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khidir HH, Eudy DM, Porras-Alfaro A, Herrera J, Natvig DO, Sinsabaugh RL. A general suite of fungal endophytes dominate the roots of two dominant grasses in a semiarid grassland. Journal of Arid Environments. 2009;74:35–42. [Google Scholar]
- 48.Knapp DG, Pintye A, Kovács GM. The dark side is not fastidious -- dark septate endophytic fungi of native and invasive plants of semi-arid sandy areas. PLoS ONE. 2012;7:e32570. doi: 10.1371/journal.pone.0032570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lackner G, Partida-Martinez LP, Hertweck C. Endofungal bacteria as producers of mycotoxins. Trends in Microbiology. 2009;17:570–576. doi: 10.1016/j.tim.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Lau MK, Arnold AE, Johnson NC. Factors influencing communities of fungal endophytes in riparian woody plants. Fungal Ecology. 2013;6:365–378. [Google Scholar]
- 51.Lopez BR, Bashan Y, Bacilio M. Endophytic bacteria of Mammillaria fraileana, an endemic rock-colonizing cactus of the southern Sonoran Desert. Archives of Microbiology. 2011;193:527–541. doi: 10.1007/s00203-011-0695-8. [DOI] [PubMed] [Google Scholar]
- 52.Loro M, Valero-Jiménez CA, Nozawa S, Márquez LM. Diversity and composition of fungal endophytes in semiarid Northwest Venezuela. Journal of Arid Environments. 2012;85:46–55. [Google Scholar]
- 53.Lucero ME, Barrow JR, Osuna P, Reyes I. Plant-fungal interactions in arid and semi-arid ecosystems: large-scale impacts from microscale processes. Journal of Arid Environments. 2006;65:276–284. [Google Scholar]
- 54.Lucero ME, Unc A, Cooke P, Dowd S, Sun S. Endophyte microbiome diversity in micropropagated Atriplex canescens and Atriplex torreyi var griffithsii. PLoS One. 2011;6:e17693. doi: 10.1371/journal.pone.0017693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lugo MA, Reinhart KO, Menoyo E, Crespo EM, Urcelay C. Plant functional traits and phylogenetic relatedness explain variation in associations with root fungal endophytes in an extreme arid environment. Mycorrhiza. 2014 doi: 10.1007/s00572-014-0592-5. [DOI] [PubMed] [Google Scholar]
- 56.Maddison DM, Maddison WP. MacClade v. 4.08a. 2009 http://macclade.org.
- 57.Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis. 2011 http://mesquiteproject.org.
- 58.Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, Abou-Hadid AF, El-Behairy UA, Sorlini C, Cherif A, Zocchi G, Daffonchio D. A drought resistance-promoting microbiome is selected by root system under desert farming. PloS One. 2012;7:e48479. doi: 10.1371/journal.pone.0048479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medina-Roldán E, Arredondo JT, Huber-Saanwald E, Chapa-Vargas L, Olalde-Portugal V. Grazing effects on fungal root symbionts and carbon and nitrogen storage in a shortgrass steppe in central Mexico. Journal of Arid Environments. 2008;72:546–556. [Google Scholar]
- 60.Moran NA. Symbiosis. Current Biology. 2006;16:R866–R871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 61.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:S8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noy-Meir I. Desert ecosystems: environment and producers. Annual Review of Ecology and Systematics. 1973;4:25–51. [Google Scholar]
- 63.Okane I, Srikitikulchai P, Tabuchi Y, Sivichai S, Nakagiri A. Recognition and characterization of four Thai xylariaceous fungi inhabiting various tropical foliages as endophytes by DNA sequences and host plant preference. Mycoscience. 2012;53:122–132. [Google Scholar]
- 64.Oono R, Lutzoni F, Arnold AE, Kaye L, U'Ren JM, May G, Carbone I. Genetic variation in horizontally transmitted symbionts of pine needles reveals population structure in cryptic species. American Journal of Botany. 2014;101:1362–1374. doi: 10.3732/ajb.1400141. [DOI] [PubMed] [Google Scholar]
- 65.Pan JJ, Baumgarten AM, May G. Effects of host plant environment and Ustilago maydis infection on the fungal endophyte community of maize (Zea mays) New Phytologist. 2008;178:147–156. doi: 10.1111/j.1469-8137.2007.02350.x. [DOI] [PubMed] [Google Scholar]
- 66.Partida-Martinez LP, Monajembashi S, Greulich KO, Hertweck C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Current Biology. 2007;17:773–777. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 67.Porras-Alfaro A, Herrera J, Sinsabaugh RL, Odenbach K, Lowrey T, Natvig DO. Novel root fungal consortium associated with a dominant desert grass. Applied and Environmental Microbiology. 2008;74:2805–2813. doi: 10.1128/AEM.02769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porras-Alfaro A, Herrera J, Natvig DO, Lipinski K, Sinsabaugh RL. Diversity and distribution of soil fungal communities in a semiarid grassland. Mycologia. 2011;103:10–21. doi: 10.3852/09-297. [DOI] [PubMed] [Google Scholar]
- 69.Porras-Alfaro A, Raghavan S, Garcia M, Sinsabaugh RL, Natvig DO, Lowrey TK. Endophytic fungal symbionts associated with gypsophilous plants. Botany. 2014;92:295–301. [Google Scholar]
- 70.Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 71.Powell BF, Halvorson WL, Schmidt CA. OFR 2007-1296. U.S. Geological Survey, Southwest Biological Science Center, Sonoran Desert Research Station, University of Arizona; Tucson, AZ: 2007. Vascular Plants and Vertebrate Inventory of Saguaro National Park, Tucson Mountain District. [Google Scholar]
- 72.Riddle JM, Arnold AE. Diversity and phylogenetic affinities of endohyphal bacteria associated with foliar fungal endophytes of the Sonoran Desert. Inoculum. 2011;62:38. [Google Scholar]
- 73.Rodriguez Estrada AE, Jonkers W, Kistler HC, May G. Interactions between Fusarium verticillioides, Ustilago maydis, and Zea mays: An endophyte, a pathogen, and their shared plant host. Fungal Genetics and Biology. 2012;49:578–587. doi: 10.1016/j.fgb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 74.Rodriguez RJ, White J, Arnold AE, Redman R. Fungal endophytes: Diversity and ecological roles. New Phytologist. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez RJ, Redman RS, Henson JM. The role of fungal symbioses in the adaptation of plants to high stress environments. Mitigation and Adaptation Strategies for Global Change. 2004;9:261–272. [Google Scholar]
- 76.Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Hickman DS, Jee J, Kimovec FM, Koppstein D, Marks DH, Mittermiller PA, Nuñez SJ, Santiago M, Townes MA, Vishnevetsky M, Williams NE, Núnez Vargas MP, Boulanger LA, Bascom-Slack C, Strobel SA. Biodegradation of polyester polyurethane by endophytic fungi. Applied and Environmental Microbiology. 2011;77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sandberg DC, Battista L, Arnold AE. Fungal endophytes of aquatic macrophytes: diverse host-generalists characterized by tissue preferences and geographic structure. Microbial Ecology. 2014;67:735–747. doi: 10.1007/s00248-013-0324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saunders M, Kohn LM. Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytologist. 2008;182:229–238. doi: 10.1111/j.1469-8137.2008.02746.x. [DOI] [PubMed] [Google Scholar]
- 79.Schoch CL, Sung GH, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh SO, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O'Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic Biology. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- 80.Strobel GA. Endophytes as sources of bioactive products. Microbes and Infection. 2003;5:535–544. doi: 10.1016/s1286-4579(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 81.Strobel GA, Knighton B, Kluck K, Ren Y, Livinghouse T, Griffin M, Spakowics D, Sears J. The production of myco-diesel hydrocarbons and their derivatives by the endophytic fungus Gliocladium roseum (NRRL 50072) Microbiology. 2008;154:3319–3328. doi: 10.1099/mic.0.2008/022186-0. [DOI] [PubMed] [Google Scholar]
- 82.Sun Y, Wang Q, Lu X, Okane I, Kakishima M. Endophytic fungal community in stems and leaves of plants from desert areas in China. Mycological Progress. 2012;11:781–790. [Google Scholar]
- 83.Suryanarayanan TS, Wittlinger SK, Faeth SH. Endophytic fungi associated with cacti in Arizona. Mycological Research. 2005;109:635–639. doi: 10.1017/s0953756205002753. [DOI] [PubMed] [Google Scholar]
- 84.Turner RM, Bowers JE, Burgess TL. Sonoran Desert Plants: an Ecological Atlas. University of Arizona Press; 2005. [Google Scholar]
- 85.U'Ren JM. Dissertation. The University of Arizona; 2011. Host-, geographic-, and ecological specificity of endophytic and endolichenic fungal communities. [Google Scholar]
- 86.U'Ren JM, Dalling JW, Gallery RE, Maddison DR, Davis EC, Gibson CM, Arnold AE. Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: a case study for analyzing fungal environmental samples. Mycological Research. 2009;113:432–449. doi: 10.1016/j.mycres.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 87.U'Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Host- and geographic structure of endophytic and endolichenic fungi at a continental scale. American Journal of Botany. 2012;99:898–914. doi: 10.3732/ajb.1100459. [DOI] [PubMed] [Google Scholar]
- 88.U'Ren JM, Riddle JM, Monacell JT, Carbone I, Miadlikowska J, Arnold AE. Tissue storage and primer selection influence pyrosequencing-based inferences of diversity and community structure of endophytic fungi. Molecular Ecology Resources. 2014;14:1032–1048. doi: 10.1111/1755-0998.12252. [DOI] [PubMed] [Google Scholar]
- 89.van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis M, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IA. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–75. [Google Scholar]
- 90.Ward D. The Biology of Deserts. Oxford: Oxford University Press; 2009. [Google Scholar]
- 91.Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends in Plant Science. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 92.Zabalgogeazcoa I, Vázquez de Aldana BR, García Ciudad A, García Criado B. Fungal endophytes in grasses from semi-arid permanent grasslands of western Spain. Grass and Forage Science. 2003;58:94–97. [Google Scholar]
- 93.Zimmerman NB, Vitousek P. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proceedings of the National Academy of Sciences USA. 2012;7:13022–13027. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 95.Zwickl DJ. Dissertation. The University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Collection details, organized by host.
Supplementary Table 2. GenBank accession numbers and ecological details for known Preussia spp. used in phylogenetic analysis.
Supplementary Table 3. Collection information, OTU designations, and BLAST matches for endophytes.
Supplementary Table 4. Summary of isolation data as a function of host species.