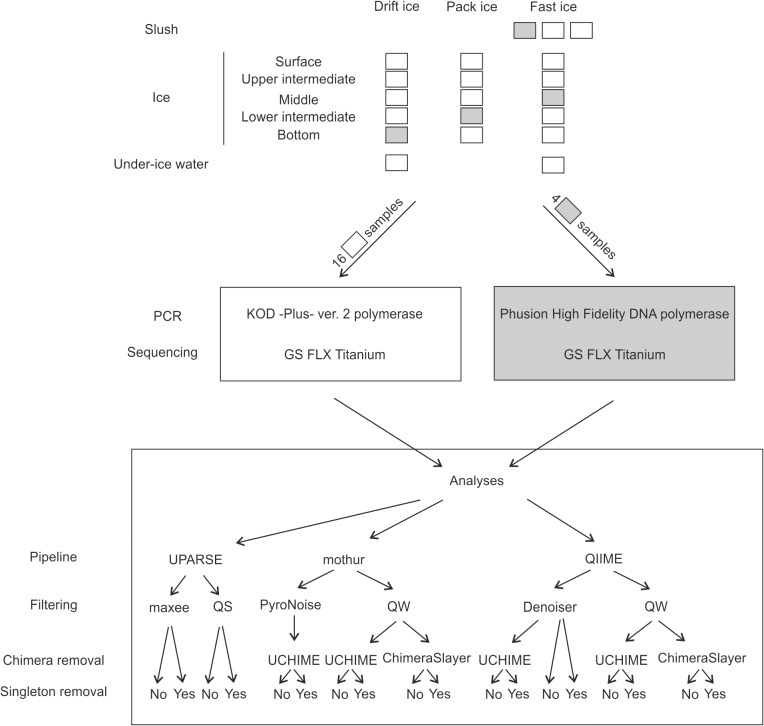
Fig 1. Experimental design.
The 20 obtained samples were divided into two sets: 16 samples were amplified and sequenced in Japan, and 4 samples (grey) were amplified and sequenced in Finland. For downstream analyses, we combined and analyzed these two sets with UPARSE, mothur and QIIME program packages. Within the program packages, we used varying quality filtering methods: in UPARSE, maximum expected error (maxee) and quality score (QS) filtering methods; in mothur, PyroNoise denoising and quality score window (QW) filtering; and in QIIME Denoiser denoising and QW filtering. In addition, we tested UCHIME and Chimera Slayer chimera detection methods in mothur and QIIME.