Figure 4.
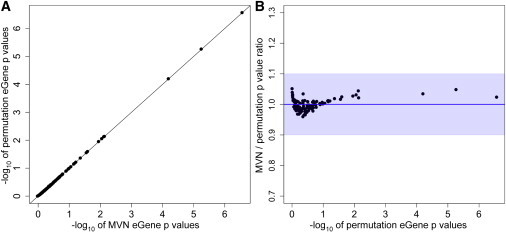
Results of Our MVN Approach Applied to 100 Probes in the Brain eQTL Dataset
We applied both distribution-reshaping and small-sample adjustment.
(A) The direct comparison of eGene p values from our MVN approach to those from the permutation test; the x axis is the −log10 of MVN eGene p values, and the y axis is the −log10 of the permutation-test eGene p values. A y = x line is drawn as a black line.
(B) The ratio of MVN eGene p values to permutation-test eGene p values (y axis) versus the −log10 of permutation-test eGene p values (x axis). A y = 1 line is drawn as a red line. The maximum ratio is 1.05, the minimum ratio is 0.96, and the average is 1. The average error rate is 1.56%, and accuracy is 98.44%. Blue shading denotes 90% accuracy.
