Figure 5.
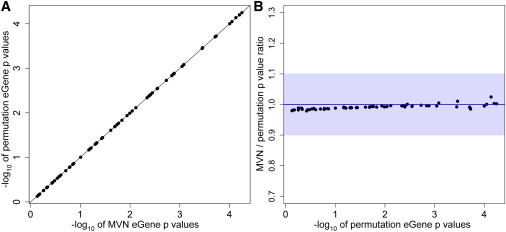
Results of the MVN Approach Applied to 100 Probes in the HapMap 3 eQTL Dataset
Because of the large sample size, we applied only the distribution-reshaping algorithm to an MVN.
(A) The direct comparison of eGene p values from our MVN approach to those from the permutation test; the x axis is the −log10 of MVN eGene p values, and the y axis is the −log10 of the permutation-test eGene p values.
(B) The ratio of MVN eGene p values to permutation-test eGene p values (y axis) versus the −log10 of the permutation-test eGene p values (x axis). The maximum ratio is 1.024, the minimum ratio is 0.979, and the average is 0.991. The average error rate is 1.0%, and accuracy is 99.0%. Blue shading denotes 90% accuracy.
