Figure 1.
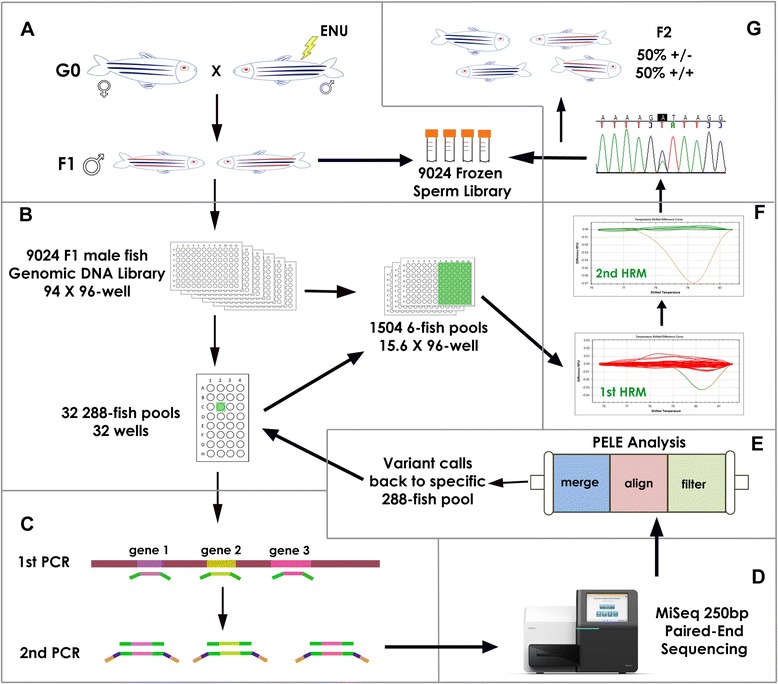
NGS-TILLING process. A: A long-term resource for many TILLING screens consisting of a genomic DNA sample and a corresponding cryopreserved sperm sample was prepared from 9,024 F1 ENU-mutagenized male zebrafish. B: Library Pooling. Normalized genomic DNA (gDNA) was pooled twice: first, gDNA from 6 fish was pooled together to make 1,504 6-fish pools in 16 96-well plates. These six-fish pools will be used for HRM identification of carrier fish (step F). Second, groups of 48 6-fish pools were pooled together into 288-fish pools (a total of 32 288-fish pools). C: Target Preparation. gDNA from 288-fish pools was used as a template for PCR amplification of ~250 bp fragments corresponding to exons of genes of interest using gene-specific primers with P5/P7 SEQ tails (green). After normalization, amplicons from each 288-fish pool were combined and used as template for a brief second PCR that added Nextera index sequences (blue) and Illumina P5/P7 sequences (yellow). D: Sequencing: All amplicons from the entire library were combined and sequenced (Illumina MiSeq platform), generating fully overlapping 250 bp paired-end sequences. E: Data Analysis. Sequence analysis using PELE and PoDATA identified rare deleterious variants (occurring in 1/100 to 1/1000 reads) in single 288-fish pools. F: Deconvolution. A fragment centered on a putative variant call was amplified from each of the 48-six-fish pools used to make up the 288-fish pool in which that variant was detected, and was subjected to High Resolution Melt (HRM) Analysis. Then HRM of the six individual fish in the six-fish pool that showed distinct melting kinetics identified the individual carrier. G: Mutant Recovery. Finally, the presence of the variant identified by PELE and PoDATA was confirmed in that fish by Sanger sequencing. F2 heterozygotes were generated by in vitro fertilization of WT eggs with the corresponding cryopreserved sperm sample.
