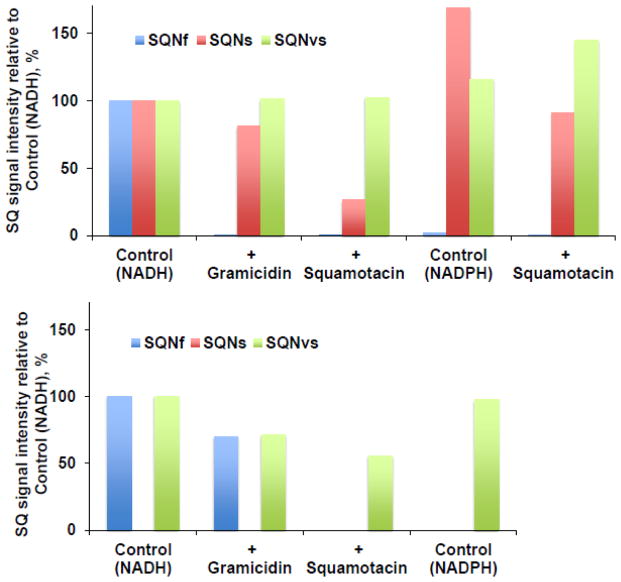
Fig. 3.
Effects of gramicidin D (an uncoupler, 100 μM), squamotacin (a potent complex I inhibitor, 100 μM), or NADPH (2 mM) on the ubisemiquinone g = 2.00 EPR signals in the wild-type complex I (A) and the ΔNuoL variant (B) reconstituted in proteoliposomes. SQNf (P1/2 = ~50 mW), SQNs (P1/2 = ~3 mW), and SQNvs (P1/2 = ~0.1 mW) were resolved by our computer fitting program. The data were obtained from the power saturation analysis at 51 mW for SQNf (blue), 3 mW for SQNs (red), and 0.08 mW for SQNvs (green). The signal amplitudes of these SQ species (peak to peak) were measured. The EPR signal intensity was normalized relative to the control (NADH). There was no SQNs (P1/2 = ~3 mW) component in the ΔNuoL variant. [WT-PL] = 1.4 mg/ml, NADH:DQ= 14.19 μmol/min/mg and NADH:Ferricyanide = 132 μmol/min/mg. [ΔNuoL-PL] = 1 mg/ml, NADH:DQ= 2.38 μmol/min/mg and NADH:Ferricyanide = 80.4 μmol/min/mg. These are the representative data from two separate sets of samples.