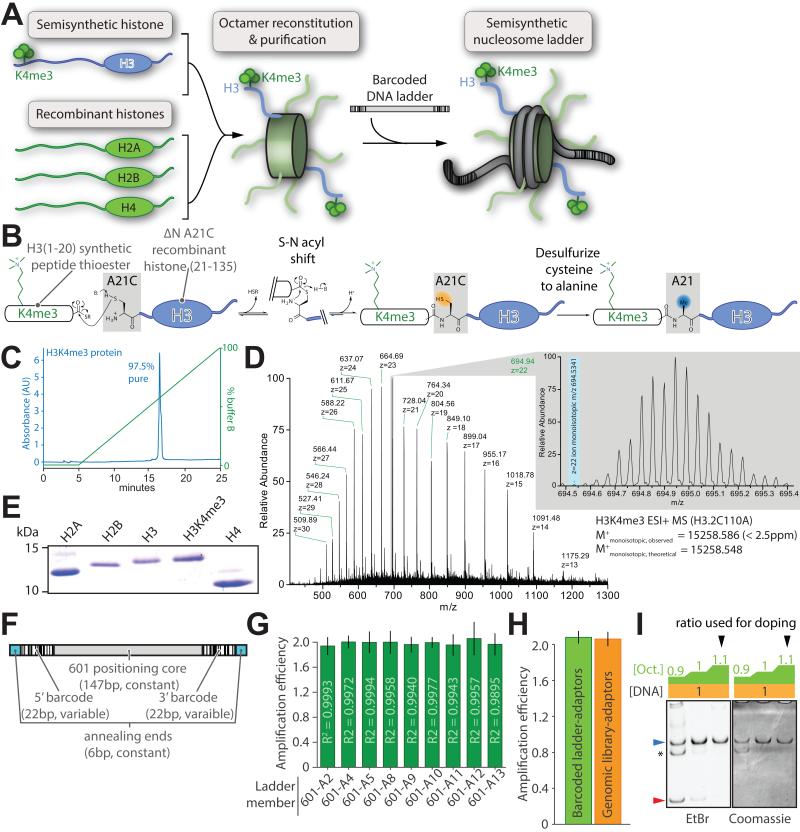
Figure 2. Design and preparation of barcoded semisynthetic nucleosomes.
(A) Schematic depiction of the reconstitution of a semisynthetic H3K4me3 nucleosome ladder: histone octamers, produced by refolding equimolar core histones from recombinant and semisynthetic sources, are purified then mixed with equal amounts of barcoded ladder DNA. (B) Semisynthesis by native chemical ligation from a K4me3-modified peptide thioester and the recombinantly produced remainder of the H3 protein bearing an N-terminal engineered cysteine. Subsequent desulfurization of this cysteine recapitulates the native alanine at this position. Characterization of the purified H3K4me3 semisynthetic histone via (C) analytical C18-HPLC and (D) electrospray mass spectrometry. (E) Representative Coomassie-stained SDS-PAGE gel of the recombinant human core histones and semisynthetic H3K4me3 used in reconstitutions. (F) Schematic representation of barcoded nucleosome positioning DNA sequences based on the 601 positioning nucleosome sequence. (G) Amplification per cycle of barcoded ladder DNA is measured with qPCR utilizing a 2× serial dilution series fit by linear regression (R2 of the fit displayed in each bar). (H) Amplification per cycle of all barcoded DNA ladder members versus native genomic DNA fragments after ligation of sequencing adaptors. (I) A representative reconstitution of internal standard nucleosome where octamer is titrated (wedge: blue, nucleosome; red, free DNA; asterisk: uncharacterized complex). All error bars are 95% CI. See also Figure S2.