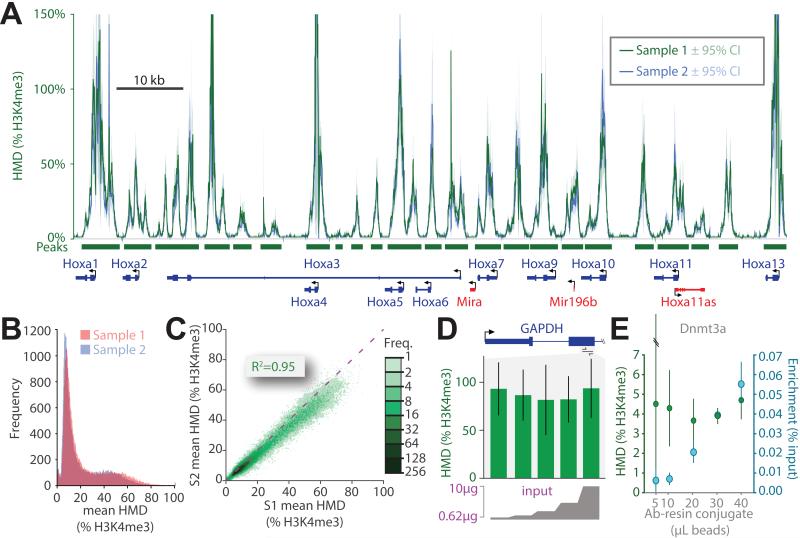
Figure 5. ICeChIP is highly reproducible and more robust to experimental differences.
(A) The superposition of HMD derived from two biological replicates (green and blue) of the H3K4me3 ICeChIP-seq in mESCs displayed at the Hoxa gene cluster. HMD for both replicates averaged over 25bp intervals and displayed with 95%CI. (B) Frequency histogram of the mean mononucleosome HMD values averaged over called peaks (p<10−20) for biological replicates. (C) Scatter plot comparison of the two samples (S1 and S2) via plotting the mean mononucleosome HMD (%H3K4me3) for called peaks; the dashed purple line indicates a slope of 1. (D) HMD measurements (%H3K4me3) are independent of input loaded into immunoprecipitation over a large interval: doped HEK293 native chromatin input (0.625-10 μg, 2 fold concentration series) analyzed by ICeChIP-qPCR at within GAPDH with 50 μL of antibody-resin conjugate. (E) Measurement of IP-enrichment and HMD (% H3K4me3) at the DNMT3a locus by ICeChIP-qPCR in mESCs with fixed 10 μg of chromatin input as a function of antibody-resin conjugate amount. See also Figure S5.