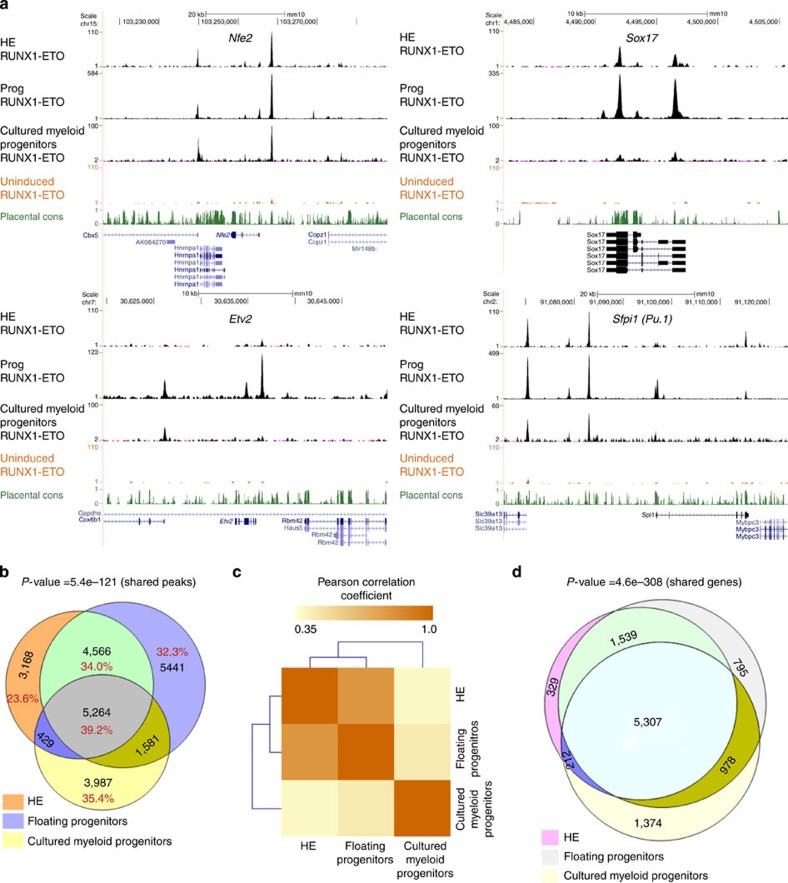
Figure 5. RUNX1-ETO binds to different binding sites but similar genes at early and late stages.
(a) UCSC genome browser screenshots of ChIP-seq data for RUNX1-ETO in HE, CD41+KIT+Tie2− progenitors and cultured myeloid progenitors at Nfe2, Sox17, Etv2 and Sfpi1 (Pu.1), (b) Venn diagram showing the percentage and number of intersections between the RUNX1-ETO ChIP-seq peaks in HE, progenitors and myeloid progenitors. The P value for calculating the significance of the overlap of all shared peaks (5,264) was obtained by bootstrapping and it was found to be highly significant, with z scores of 23.36 (P value<5.45e−121). (c) Pearson correlation analysis of the ChIP-seq data obtained with HE, progenitors and myeloid progenitors demonstrating that HE and progenitors from the blast culture cluster away from the myeloid progenitors, which have progressed further in differentiation. Pearson's correlation coefficients were calculated between all union peaks using the normalized read counts. A correlation matrix was generated and Pearson correlation coefficients are displayed after hierarchical clustering as a heat map. (d) Venn diagram showing the overlap of RUNX1-ETO bound genes in HE, progenitors and cultured progenitors. The shared gene overlap was found to be significant with P value<4.6e−30 (ref. 8).