Fig. 5.
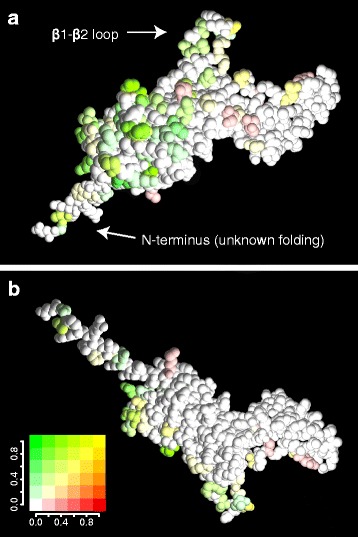
Mapping variable amino acid residues on the PsbO structure. a View from thylakoid lumen, (b) view from PSII. Differences between isoforms (in green) are merged with differences between species (in red); the frequency of particular differences on each position is indicated by colour gradient. The homology model of the Solanum tuberosum PsbO2 based on the X-ray structure of cyanobacterial PsbO [PDB:3ARC] [5] was constructed using Swiss-Model program [38]; the first 13 N-terminal amino acids were not present in the template structure, so they were pasted in the model without attempts to show any folding
