Figure 2.
RNA-Seq Reveals Unique Transcriptional States of hPGCs
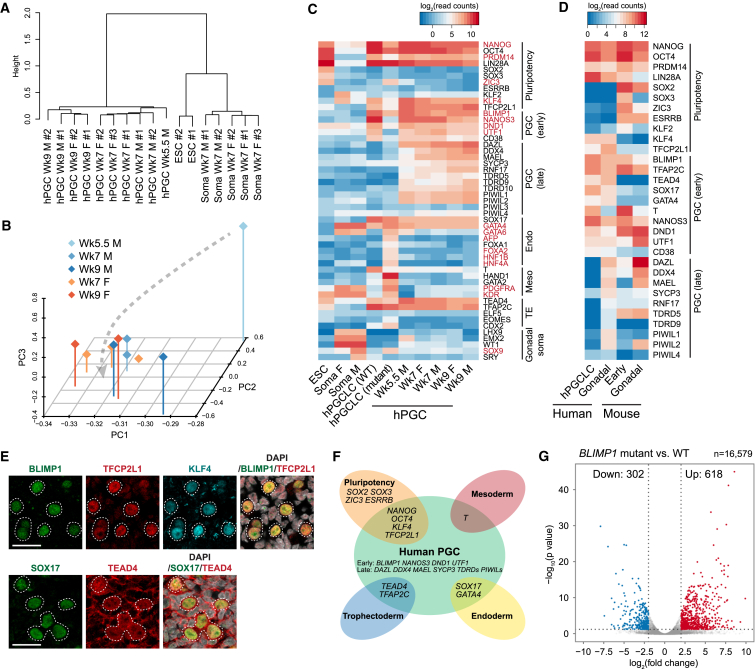
(A) Hierarchical clustering of gene expression profiles. Biological replicates of Wk5.5–Wk9 male (M) and female (F) hPGCs, gonadal somatic cells (soma), and conventional H9 ESCs are shown. Note that only one Wk5.5 hPGC sample was available for RNA-seq.
(B) Principal component analysis (PCA) of gene expression in hPGC samples. Arrow line indicates developmental progression along PC2 and PC1.
(C) Heatmap showing mean expression of representative genes in human samples. Differentially expressed genes between day 4 wild-type (WT) and BLIMP1 mutant hPGCLCs [log2(fold change)>2, p < 0.05] are highlighted. Note that mutant cells lack BLIMP1 protein as determined by immunofluorescence, but frame-shifted mutant transcripts are detected. “Endo,” endoderm; “Meso,” mesoderm; “TE,” trophectoderm.
(D) Expression of key genes in human and mouse PGCs. Mean expression in biological replicates of hPGCLCs, Wk7–Wk9 hPGCs (gonadal), E7.5 (early), and E11.5–12.5 (gonadal) mPGCs are shown.
(E) Immunofluorescence of TFCP2L1, KLF4, and TEAD4 on Wk7 female genital ridge cryosections. hPGCs are counterstained by BLIMP1 or SOX17. Scale bars, 20 μm.
(F) Schematic illustrating the unique transcriptome of hPGCs.
(G) Volcano plot showing differentially expressed genes between day 4 BLIMP1 mutant and wild-type (WT) hPGCLCs [log2(fold change)>2, p < 0.05].