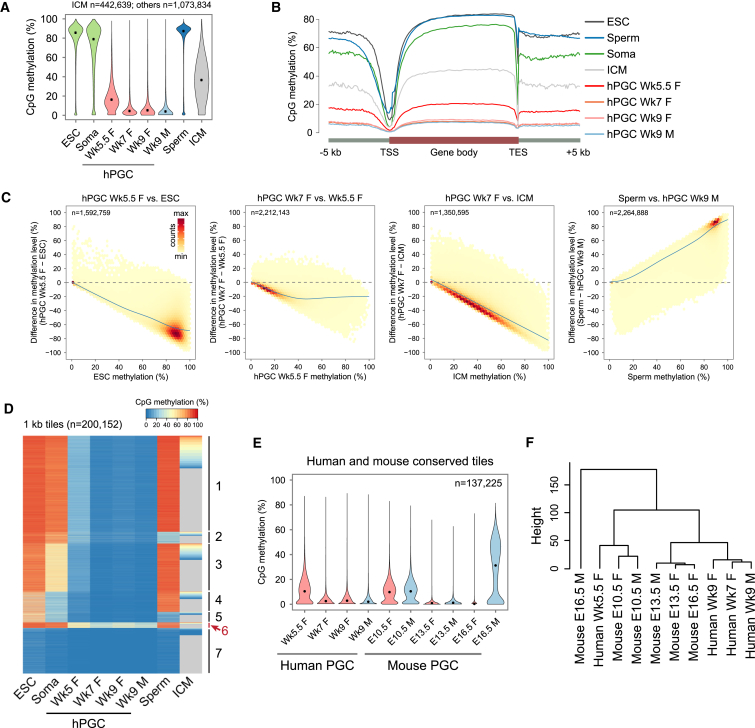
Figure 4.
Comprehensive DNA Demethylation in hPGCs Revealed by Base-Resolution BS-Seq
(A) Violin plots showing distribution of CpG methylation levels in overlapped 1 kb genomic tiles of conventional H9 ESCs, Wk7 female gonadal somatic cells (soma), Wk5.5–Wk9 female and male hPGCs, sperm, and ICM. Common tiles with a minimum of 5 CpGs and at least 20% of the total CpGs covered by at least 5× in each sample are considered. These thresholds were applied to all subsequent methylation analyses unless stated otherwise. Due to low coverage, ICM only has ∼42% of common tiles fulfilling the above criteria. Black point indicates median.
(B) Averaged CpG methylation level profiles of all genes from 5 kb upstream (−) of transcription start sites (TSSs), through scaled gene bodies to 5 kb downstream (+) of transcription end sites (TES).
(C) Density plots illustrating DNA methylation dynamics of 1 kb tiles between indicated pairs of samples. Color intensity indicates tile counts within each bin, whereas blue regression lines show trends of methylation changes.
(D) K-means clustering of repeat-free tiles into seven dynamic groups. Gray tiles of ICM do not pass coverage thresholds and are therefore not shown. Tiles in cluster 6 retain partial methylation in hPGCs.
(E) Violin plots showing distribution of CpG methylation levels in human and mouse PGCs at conserved repeat-free 1 kb tiles.
(F) Unsupervised hierarchical clustering of methylation levels of conserved repeat-free 1 kb tiles in human and mouse PGCs.