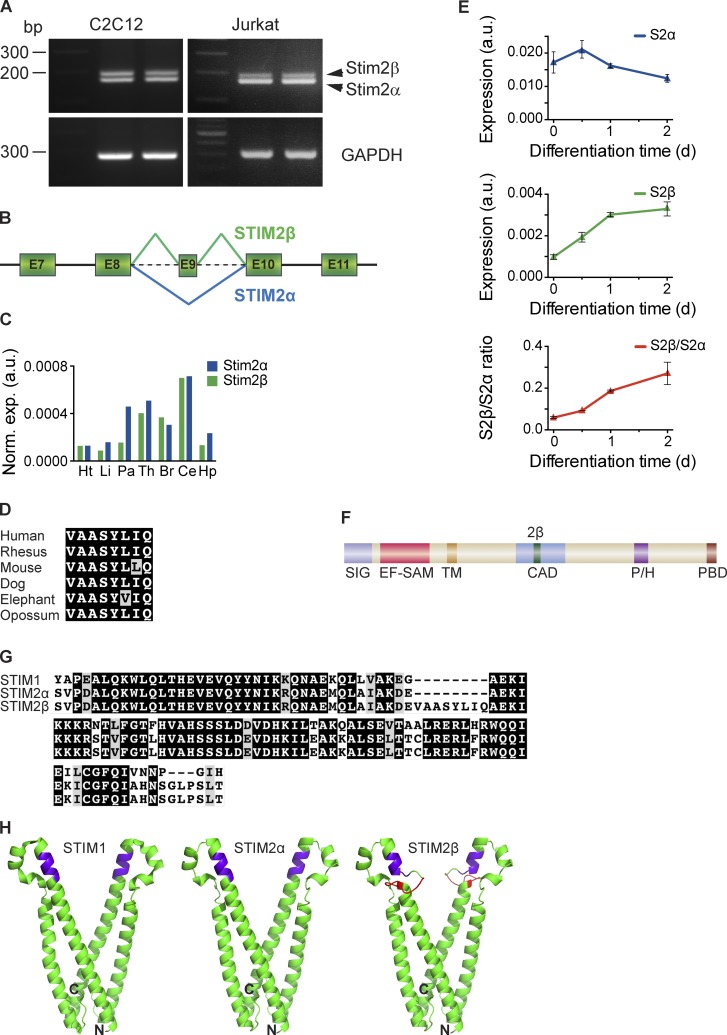
Figure 1.
STIM2β is a novel, widely expressed STIM2 splice isoform. (A, top) cDNA from Jurkat and C2C12 cells was amplified using primers targeting the CAD domain of STIM2. The topmost band represents the STIM2β splice isoform. (bottom) Primers targeting GAPDH were used as a positive control. (B) Partial schematic of the STIM2 genomic locus. In-frame inclusion of exon 9 produces STIM2β. (C) GAPDH-normalized expression levels of STIM2α and STIM2β in human tissue RNA samples, measured by quantitative RT-PCR. Means of technical replicates are shown (Ht, heart; Li, liver; Pa, pancreas; Th, thymus; Br, brain; Ce, cerebellum; Hp, hippocampus). (D) Sequence alignment of the 2β insert across six mammalian species (also see Fig. S1 A). Conservative differences are marked in gray. (E) GAPDH-normalized expression levels of STIM2α (top) and STIM2β (middle) mRNA in differentiating cultured C2C12 myoblasts. An approximately fivefold increase in the STIM2β/STIM2α ratio (bottom) occurs during the first 2 d of differentiation. Error bars represent SEM of three independent wells. (F) Domain structure of STIM2. Inclusion of exon 9 leads to an insert (2β, green) in the CAD (SIG, signal peptide; EF-SAM, EF hand/sterile-α motif; TM, transmembrane segment; P/H, proline/histidine-rich domain; PBD, polybasic domain). (G) Alignment of partial CAD sequences from human STIM1, STIM2α, and STIM2β. Sequence identity and similarity are shown in black and gray, respectively. (H) Predicted structures of STIM2α- (center) and STIM2β-CAD (right) derived from the crystal structure of STIM1-CAD (left; Yang et al., 2012) by homology modeling. The stretch of basic residues involved in Orai1 binding is highlighted in purple, and the 2β insert is highlighted in red. N and C termini of the front monomer in each structure are marked for orientation. a.u., arbitrary unit.