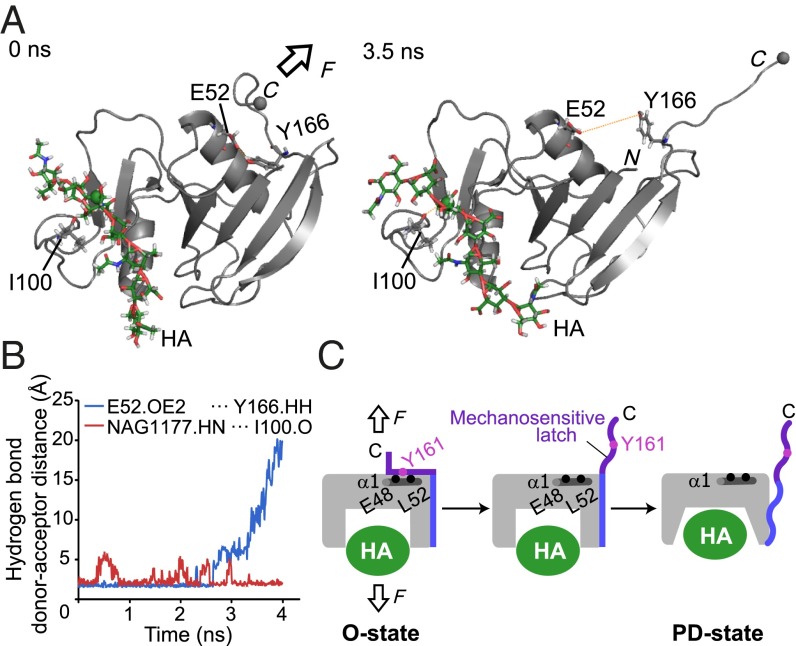
Fig. 4.
Steered molecular dynamics (SMD) simulation of CD44 HABD by pulling force. (A) Snapshots of the HABD–HA8 complex at 0 ns (Left) and 3.5 ns (Right) during the SMD simulation. In the SMD simulation, the Cα carbon of the C-terminal residue Ile173 (gray sphere) was pulled at 10 Å/ns in the indicated direction (arrow), whereas the C2 atom of the N-acetylglucosamine residue (green sphere) was kept fixed. In the snapshot after 3.5 ns in the SMD simulation, the C-terminal mechanosensitive latch was completely separated from the α1 helix. (B) The time course of the hydrogen bond donor–acceptor distances between HA NAG1177 and I100 (red) and E52 and Y166 (blue). (C) Schematic depiction of the mechanosensitive latch in the force-induced conformational change of CD44 HABD.