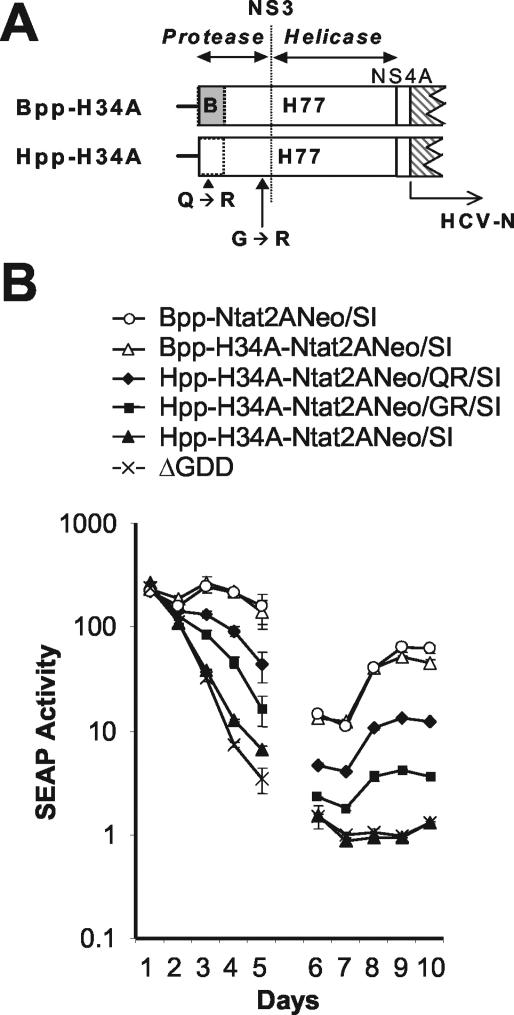
FIG. 3.
(A) Schematic depicting the organization of the 5′ end of the second open reading frame in subgenomic chimeric replicons containing most (Bpp-H34A-Ntat2ANeo/SI) or all (Hpp-H34A-Ntat2ANeo/SI) of the H77 genotype 1a NS34A-coding sequence in the background of the genotype 1b Bpp-Ntat2ANeo/SI. Genotype 1a sequence (H77) is shown as an open box, genotype 1b sequence (Con1 or HCV-N) as a shaded box. “Bpp” indicates the presence of genotype 1b sequence from the Con1 strain of HCV in the 5′-proximal protease coding sequence, whereas “Hpp” indicates that this sequence is derived from the genotype 1a H77 sequence. Approximate locations are shown for the adaptive mutations Q1067R and G1188R, identified in G418-resistant cell clones selected following transfection of Hpp-H34A-Ntat2ANeo/SI. The dotted line indicates the approximate border of the NS3 protease and helicase domains (between residues 1206 and 1207). (B) SEAP activity present in supernatant culture fluids collected at 24 h intervals following transfection of En5-3 cells with various chimeric 1a-1b replicons including Bpp-H34A-Ntat2ANeo/SI, Hpp-H34A-Ntat2ANeo/SI, Hpp-H34A-Ntat2ANeo/QR/SI, and Hpp-H34A-Ntat2ANeo/GR/SI. Control cells were transfected with Bpp-Ntat2ANeo/SI and the replication-defective ΔGDD mutant. See legend to Fig. 2 for further details. See Table 1 for identification of adaptive mutations in each transcript.