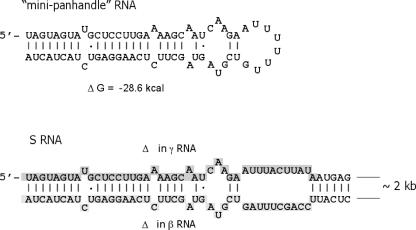
FIG. 4.
Viral panhandle RNA. At the top is shown the secondary structure of minipanhandle RNA used as a binding substrate for the SNV N-protein interaction. Mfold was used to analyze multiple alternative RNA secondary structures. The structure shown is the most probable on the basis of thermodynamic considerations. At the bottom is the terminal structure likely to be present in full-length S-segment RNA and in α RNA. The same structure formed by the terminal nucleotides in the S RNA and α RNA is shown at the bottom. Analysis of 20 alternative theoretical secondary structures indicated that the panhandle is likely to form in each. In addition, the shaded nucleotides are those deleted in γ or β RNA. The shaded nucleotides near the 5′ end are deleted in γ RNA, and those in the 3′ end are deleted in β RNA.