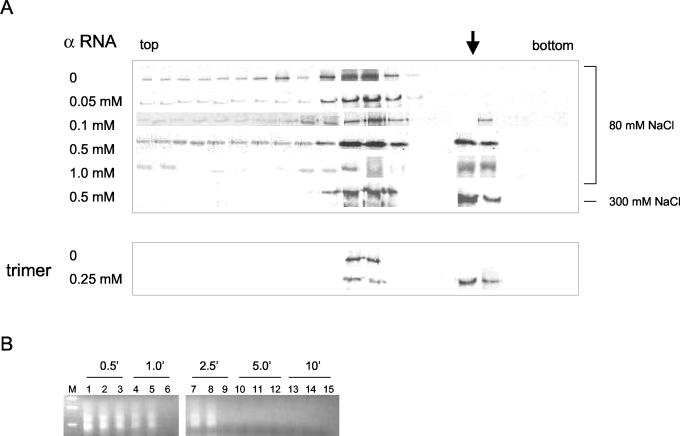
FIG. 5.
Sucrose gradient analysis of N-protein-viral RNA complexes. (A) Following sedimentation through sucrose gradients N protein was detected by Western analysis with anti-N antibody as in Fig. 2D. In gradients 1 through 6, 50 μM unfractionated N was incubated with increasing amounts of α RNA for 2 h at room temperature prior to sedimentation as indicated. All samples were incubated prior to and during centrifugation in 80 mM NaCl, except the sample labeled 300 mM, for which both the binding and sedimentation were carried out in the presence of 300 mM NaCl. In the last two gradients, 25 μM purified trimeric N protein was incubated in the absence or presence of 0.25 μM α RNA for 2 h at room temperature prior to sedimentation and Western analysis. The arrow indicates the position of the N-RNA complexes. (B) The material from fractions 5 and 6, which contained putative N-α RNA complexes, were treated with RNases A and T1 for various times (as indicated), and remaining undegraded α RNA was detected by RT-PCR and electrophoresis on a DNA gel as described in the text. Lanes: M, DNA size markers; 1, 4, 7, 10, and 13, material from fraction 5; 2, 5, 8, 11, and 14, material from fraction 6; 3, 6, 9, 12, and 15, α RNA in sucrose gradient buffer plus 20% sucrose in the absence of N protein.