Table 2. Data collection and processing.
Values in parentheses are for the outer shell.
| Data set | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Protein sample | cgAUS1-a1 | cgAUS1-a2 | cgAUS1-a2 | cgAUS1-ln | cgAUS1-lr | cgAUS1-lr | cgAUS1-lr |
| Crystallization condition | A | A | B | C | D | E, native | F, soaked in H2O2 |
| Diffraction source | P14, EMBL, DESY | I04-1, Diamond | P11, DESY | ID23, ESRF | P11, DESY | P11, DESY | I04-1, Diamond |
| Wavelength () | 1.23953 | 0.9173 | 1.0247 | 0.972499 | 1.0247 | 1.0247 | 0.9173 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Detector | PILATUS 6M-F | PILATUS 2M | PILATUS 6M | PILATUS 6M | PILATUS 6M | PILATUS 6M | PILATUS 2M |
| Crystal-to-detector distance (mm) | 223.63 | 158.8 | 301.87 | 386.862 | 301.87 | 277.77 | 236.19 |
| Rotation range per image () | 0.1 | 0.4 | 0.2 | 0.1, helical | 0.2 | 0.2 | 0.3 |
| Total rotation range () | 180 | 180 | 210 | 180 | 200 | 240 | 280 |
| Exposure time per image (s) | 0.2 | 0.4 | 0.2 | 0.043 | 0.4 | 0.2 | 0.2 |
| Space group | P212121 | P1211 | P3121 | P1211 | P1 | P1211 | P1211 |
| a, b, c () | 88.72, 90.56, 182.59 | 51.53, 183.52, 78.09 | 137.10 137.10 209.52 | 62.57, 174.11, 102.54 | 62.85, 103.79, 175.56 | 52.99, 110.41, 94.99 | 52.88, 109.75, 94.76 |
| , , () | 90.0, 90.0, 90.0 | 90.0, 94.50, 90.0 | 90.0, 90.0, 120.0 | 90.0, 105.268, 90.0 | 100.686, 91.899, 105.182 | 90, 95.761, 90 | 90, 96.297, 90 |
| Mosaicity () | 0.209 | 0.197 | 0.075 | 0.203 | 0.268 | 0.175 | 0.187 |
| Resolution range () | 45.651.62 (1.681.62) | 48.101.64 (1.701.64) | 48.921.93 (2.001.93) | 48.772.50 (2.592.50) | 47.912.93 (3.032.93) | 48.142.08 (2.152.08) | 47.411.93 (2.001.93) |
| Total No. of reflections | 1194470 (90878) | 599586 (62008) | 2019106 (201888) | 242807 (24274) | 176284 (16772) | 291243 (27660) | 305134 (30573) |
| No. of unique reflections | 186036 (18199) | 173601 (17565) | 170779 (16875) | 71373 (7074) | 88742 (8838) | 64197 (6288) | 79591 (7941) |
| Completeness (%) | 99.64 (98.42) | 98.64 (99.65) | 99.99 (99.98) | 97.84 (97.68) | 98.49 (97.9) | 98.44 (97.56) | 98.67 (99.11) |
| Multiplicity | 6.42 (4.99) | 3.45 (3.53) | 11.82 (11.96) | 3.4 (3.43) | 1.99 (1.90) | 4.54 (4.40) | 3.83 (3.85) |
| I/(I) | 11.6 (2.0) | 8.6 (2.0) | 12.2 (2.0) | 7.6 (2.0) | 4.9 (2.0) | 11.4 (2.0) | 10.7 (2.0) |
| R p.i.m. † | 0.045 (0.397) | 0.065 (0.426) | 0.043 (0.463) | 0.093 (0.545) | 0.091 (0.417) | 0.051 (0.454) | 0.056 (0.389) |
| CC1/2 ‡ | 0.998 (0.693) | 0.993 (0.604) | 0.999 (0.676) | 0.986 (0.563) | 0.979 (0.691) | 0.996 (0.765) | 0.996 (0.754) |
| Overall B factor from Wilson plot (2) | 14.88 | 16.04 | 28.18 | 47.54 | 48.28 | 36.49 | 26.11 |
| No. of molecules per asymmetric unit | 4 | 4 | 6 | 4 | 8 | 2 | 2 |
| Matthews coefficient (3Da1) | 2.20 | 2.21 | 2.28 | 2.29 | 2.30 | 2.35 | 2.32 |
| Solvent content (%) | 44.23 | 44.43 | 46.02 | 46.25 | 46.47 | 47.62 | 47.02 |
R
p.i.m. = 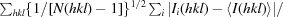
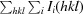 , where Ii(hkl) is the ith observation of reflection hkl and I(hkl) is the weighted average intensity for all observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and I(hkl) is the weighted average intensity for all observations of reflection hkl.
CC1/2 is defined as the correlation coefficient between two random half data sets, as described by Karplus Diederichs (2012 ▶).
