Table 1. Crystal data for the antifolates under study.
Values in parentheses are for the highest resolution shell.
| hDHFR22NADPH | hDHFR26NADPH | pcDHFR24NADPH | |
|---|---|---|---|
| PDB code | 4qhv | 4qjc | 4qjz |
| Data collection | |||
| Space group | H3 | H3 | P21 |
| Unit-cell parameters | |||
| a () | 84.4 | 84.5 | 36.7 |
| b () | 84.4 | 84.5 | 42.9 |
| c () | 78.3 | 77.9 | 61.0 |
| () | 90.0 | 90.0 | 90.0 |
| () | 90.0 | 90.0 | 95.3 |
| () | 120.0 | 120.0 | 90.0 |
| Beamline | 11-1, SSRL | 11-1, SSRL | 11-1, SSRL |
| Resolution () | 34.51.54 (1.621.54) | 34.41.54 (1.621.54) | 35.01.61 (1.721.61) |
| Wavelength () | 0.975 | 0.975 | 0.975 |
| R merge † (%) | 0.026 (0.79) | 0.062 (0.75) | 0.050 (0.70) |
| Completeness (%) | 92.8 (75.4) | 96.3 (83.0) | 95.8 (97.0) |
| Observed reflections | 72513 (7697) | 97495 (9939) | 61131 (9036) |
| Unique reflections | 28762 (3398) | 29910 (3757) | 23617 (3467) |
| I/(I) | 21.3 (4.5) | 10.2 (1.2) | 12.6 (1.5) |
| Multiplicity | 2.5 (2.3) | 3.3 (2.6) | 2.6 (2.6) |
| Refinement model | |||
| Resolution () | 34.51.54 | 34.41.54 | 35.01.61 |
| No. of reflections | 24506 | 24614 | 22409 |
| R factor‡ | 0.178 | 0.183 | 0.204 |
| R free § | 0.222 | 0.217 | 0.244 |
| Total protein atoms | 1757 | 1678 | 1851 |
| Total waters | 154 | 105 | 73 |
| Average B factor (2) | 19.1 | 23.2 | 22.3 |
| R.m.s.d. from ideal | |||
| Bond lengths () | 0.025 | 0.026 | 0.02 |
| Bond angles () | 2.85 | 2.52 | 2.19 |
| Luzzati plot error () | 0.168 | 0.218 | 0.205 |
| Ramachandran plot (%) | |||
| Most favored | 98.4 | 98.4 | 95.5 |
| Additional allowed | 1.1 | 1.6 | 3.0 |
| Generously allowed | 0.5 | 0.0 | 1.5 |
| Disallowed | 0.0 | 0.0 | 0.0 |
R
merge = 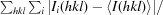
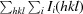 , where I(hkl) is the mean intensity of a set of equivalent reflections.
, where I(hkl) is the mean intensity of a set of equivalent reflections.
R factor = 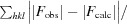
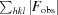 , where F
obs and F
calc are observed and calculated structure-factor amplitudes, respectively.
, where F
obs and F
calc are observed and calculated structure-factor amplitudes, respectively.
R free was calculated as for the R factor for a random 5% subset of all reflections.
