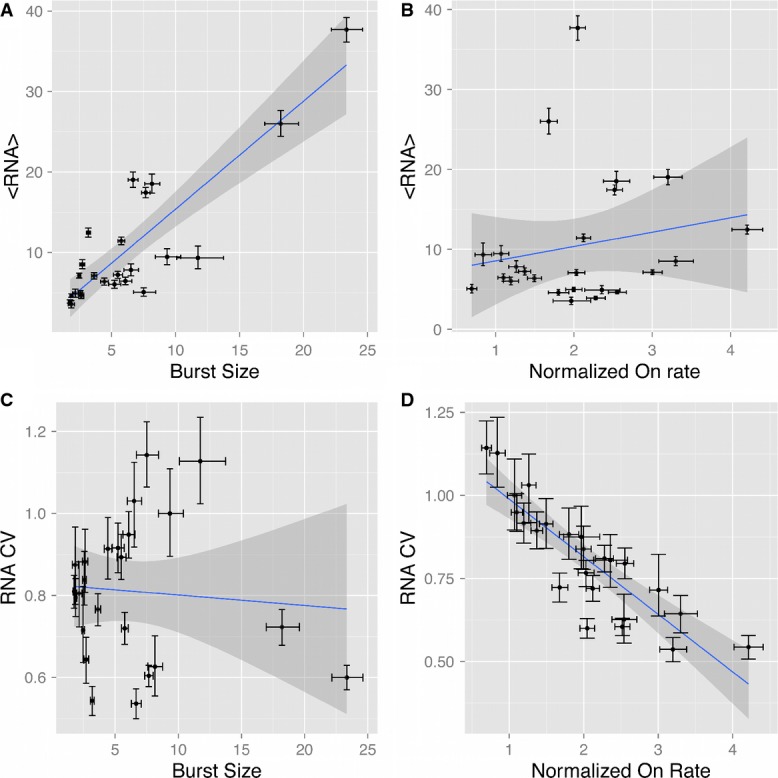
Maximum-likelihood fitting of a stochastic model of transcription to smFISH distributions reveals burst size primarily accounts for expression mean while promoter transitions from Off to On primarily accounts for expression noise
Burst size accounts for variation in mean expression. Maximum-likelihood estimation of burst size for each clone significantly accounts for a majority of the variation observed in mean RNA copy number across integration positions (slope = 1.35 ± 0.35, R2 = 0.75, rs = 0.8, P < 0.001).
Promoter On transitions are uncorrelated from expression mean. Maximum-likelihood estimation of promoter On rate normalized by the rate RNA degradation cannot account for variation in mean RNA copy number (R2 = 0.05, rs = 0.2, P = 0.37).
Burst size is uncorrelated from expression noise. Burst size cannot account for the variation in observed expression noise (CV) (R2 = 0.01, rs = 0.01).
Promoter On transitions accounts for expression noise variation. Normalized On rate (by rate of RNA degradation) significantly accounts for variation in expression noise (CV) across integration positions (slope = −0.52 ± 0.14, R2 = 0.75, rs = −0.89, P < 0.001). This independence in cross-correlations suggests that while both burst size and promoter On rate vary across integration positions, expression mean and noise are controlled by primarily orthogonal mechanisms.
Data information: Error bars on RNA moments represent 95% confidence interval estimates derived from bootstrapped smFISH distributions for each clone. Error bars on model parameters represent 95% confidence intervals estimated using 1.92 log-likelihood ratio units.
rs represents the Spearman correlation coefficient for the explanatory and response variables in each pairwise regression and
P-values represent support for correlation.