Figure 6.
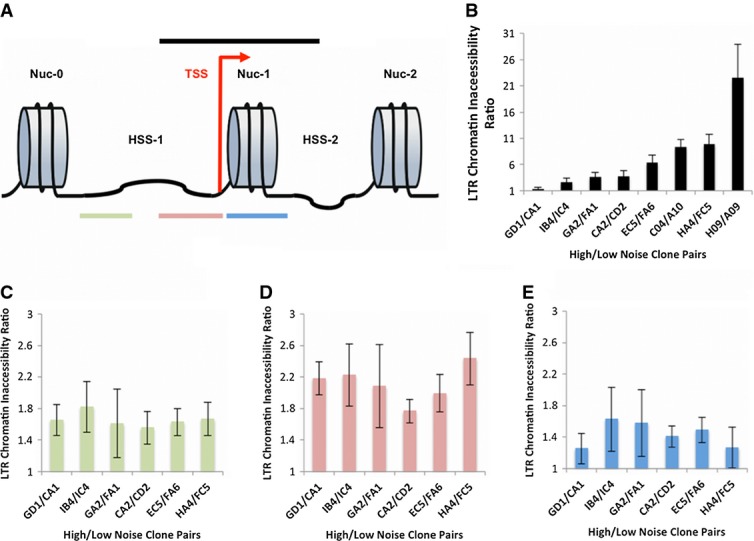
- A HIV-1 LTR exhibits positioned nucleosomes. Schematic figure showing the well-characterized placement of nucleosomes along the HIV-1 promoter. Nuc-1 is positioned at the transcription start site (TSS), and Nuc-0 is placed further upstream along the promoter.
- B Significant differences in DNA accessibility around TSS between high and low noise clones. The region highlighted in black was probed after digestion with DNase I for 16 clones. The figure plots the level of chromatin inaccessibility for pairs of clones that exhibit similar mean levels of expression. Clones that exhibit noisier gene expression also have more closed chromatin.
- C–E Differential DNA accessibility between high and low noise clones across three distinct sites in the LTR. The HIV-1 promoter was probed in greater detail with the color scheme matching that shown in (A). As in (B), it was found that for similar mean levels of gene expression, noisier clones exhibited more closed chromatin along the entire length of the promoter (ratios > 1 in all cases). Interestingly, compared to the two other regions of the promoter, the hypersensitive site (HSS) showed maximum differences in chromatin inaccessibility between high and low noise clones.
