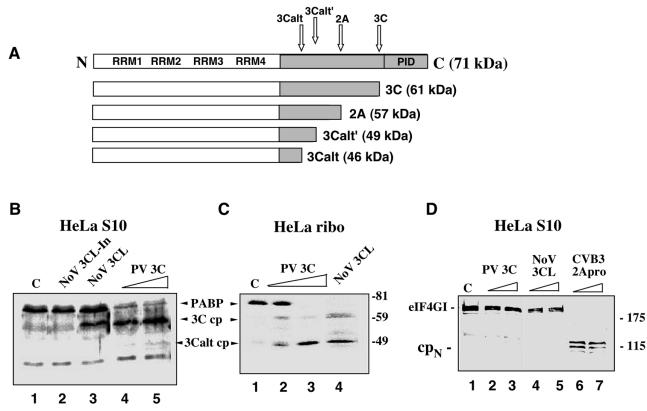
FIG. 1.
Comparison of the cleavage activity of NoV r3CLpro with that of PV 3Cpro and 2Apro on the cellular protein substrates, PABP and eIF4G. (A) The structural organization of human PABP and the previously mapped cleavage sites recognized by the PV 3C and 2A proteinases (32, 41). (B and C) NoV r3CLpro cleaves PABP. HeLa S3 cells were fractionated into cytoplasmic (S10) (B) and ribosome-associated fractions (C). In vitro cleavage reactions were assembled in these fractions and proteins were analyzed by SDS-10% PAGE followed by immunoblotting with PABP-specific antibodies. Numbers on the right show the migration of molecular weight markers (in thousands). HeLa S10 lysates (B) were incubated for 3.5 h at 37°C with the following: lane 1, mock incubation control (indicated by C); lane 2, heat-inactivated NoV proteinase, 3CL-In; lane 3, NoV r3CLpro, 80 ng/μl; lane 4, PV r3Cpro, 40 ng/μl; and lane 5, PV r3Cpro, 80 ng/μl. The HeLa ribosomal fraction (C) was incubated for 3.5 h at 37°C with the following: lane 1, mock control (indicated by C); lane 2, PV r3Cpro, 40 ng/μl; lane 3, PV r3Cpro, 80 ng/μl; and lane 4, NoV r3CLpro, 80 ng/μl. (D) NoV 3CLpro does not cleave eIF4G. Prior to immunoblot analysis with eIF4G antibody, HeLa S10 lysates were incubated for 3.5 h at 37°C with the following: lane 1, mock control (indicated by C); lane 2, PV r3Cpro, 40 ng/μl; lane 3, PV r3Cpro, 80 ng/μl; lane 4, NoV r3CLpro, 40 ng/μl; lane 5, NoV r3CLpro, 80 ng/μl; lane 6, CVB3 r2Apro, 40 ng/μl; and lane 7, CVB3 r2Apro, 80 ng/μl. The 115-kDa N-terminal cleavage fragment of eIF4G is indicated as cpN.