Fig. 1. Workflow to identify and quantify the relative abundance of CrkL-SH3 binding proteins in murine embryonic brain and liver extracts.
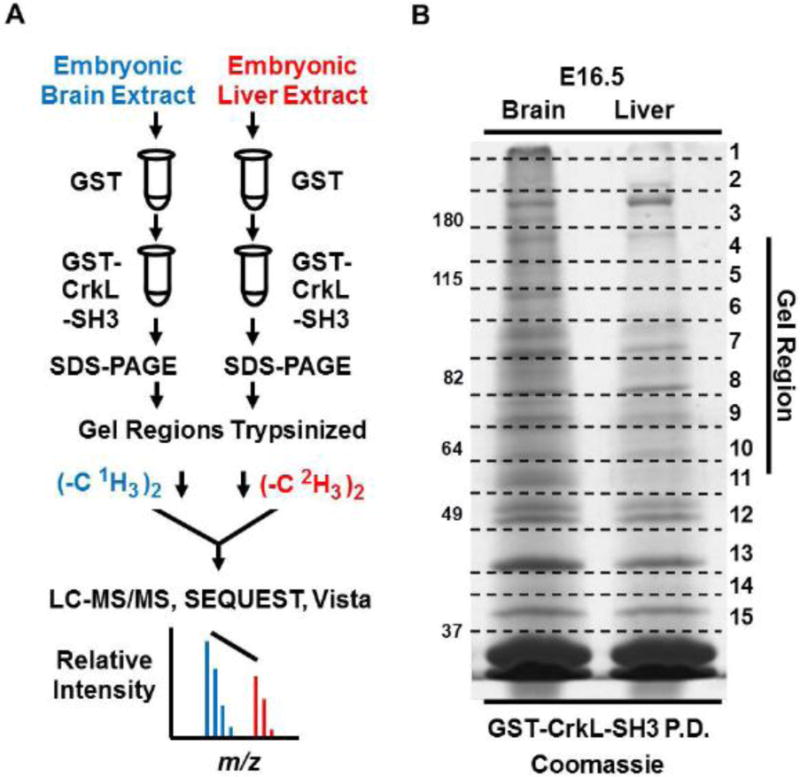
(A) Tissue extracts were pre-cleared with GST resin in batch format. The supernatants were then mixed with GST-CrkL-SH3 resin. Proteins in the pulldowns were subjected to SDS-PAGE. Gel regions were subjected to in-gel digestion and peptides originating from embryonic brain and liver were dimethylated using reagents without or with deuterium respectively. Peptides from the two samples were combined, desalted and subjected to LC-MS/MS. MS data were searched using SEQUEST, and MS1-based relative peptide quantification was performed using Vista. (B) Coomassie-stained gel of experiment one showing GST-CrkL-SH3pulldowns from embryonic murine brain and liver. Gel regions cut for in-gel tryptic digestion are indicated.
