Figure 4. ERG binds to TEAD4-interacting regions of chromatin and transactivates Hippo pathway target genes in immortalized prostate epithelial cells.
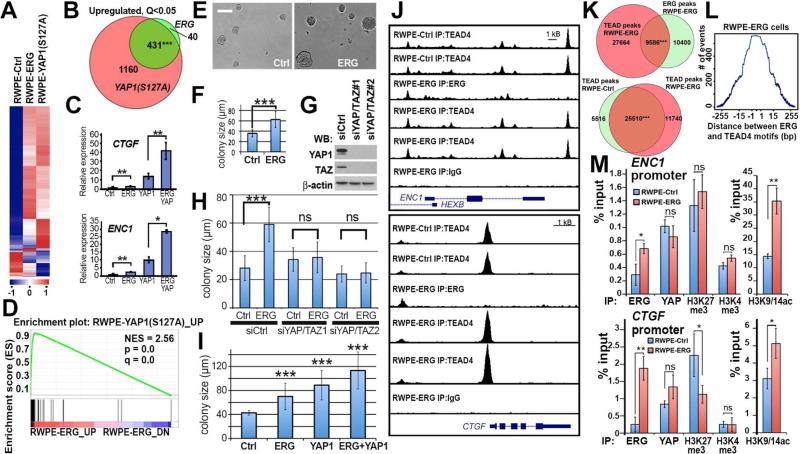
(A-B) Significant overlap of gene expression changes induced by overexpression of ERG and constitutively active YAP1. Gene expression in RWPE-1 cells transduced with empty vectors (RWPE-Ctrl), ERG (RWPE-ERG) or YAP1S127A (RWPE-YAP1S127A) was analyzed by RNASeq. n=2 per each genotype. (A) Heat-map; (B) overlap between significantly (Q<0.05) upregulated genes. Two tailed Chi-square with Yates correction.
(C) qRT-PCR analyses of two Hippo pathway target genes CTGF and ENC1 in RWPE-Ctrl (Ctrl), RWPE-ERG (ERG), RWPE-YAP1S127A (YAP1) and RWPE-ERG and YAP127A (ERG YAP) expressing RWPE-1 cells. Data represent mean ± standard deviation (n = 3) from one of three independent experiments. Student's t test.
(D) GSEA analysis of similarities between ERG-mediated gene expression changes and YAP1S127A upregulated genes.
(E-F) Brightfield images (E) and size quantitation (F) of RWPE-Ctrl (Ctrl) and RWPE-ERG (ERG) cell colonies formed after 6 days in 3D organoid culture system. The graph shows mean ±SD. n≥40. Student's t-test. Bar correspond to 100 μm.
(G) Western blot (WB) analyses of RWPE-1 cells transfected with indicated siRNA oligos and analyzed with indicated antibodies.
(H) Colony size quantitation of RWPE-Ctrl (Ctrl) and RWPE-ERG (ERG) cells in 3D organoid culture system transfected with indicated siRNA oligos. The graph shows mean ±SD. n≥60. Student's t-test.
(I) Colony size quantitation of RWPE-Ctrl (Ctrl), RWPE-ERG (ERG), RWPE-YAP1S127A (YAP1) and RWPE-ERG + YAP127A (ERG + YAP1) cells in 3D organoid culture system. The graph shows mean ±SD. n≥44. Student's t-test.
(J) UCSC genome browser views of ChIP-Seq data from RWPE-Ctrl and RWPE-ERG cells using IgG, anti-ERG and anti-TEAD4 antibodies.
(K) Highly significant overlap between positions of TEAD4 and ERG peaks in RWPE-ERG cells (hypergeometric p-value=3.761525e-263 calculated by Bioconductor package “ChIPPeakAnno”). (L) Tendency toward close juxtaposition of ERG (A/C)GGAA(G/A) and TEAD4 GGAAT(G/T)(T/C) recognition motifs in ERG-TEAD4 overlapping peaks in RWPE-ERG cells.
(M) qPCR analysis of ChIP experiments from RWPE-Ctrl and RWPE-ERG cells using indicated antibodies. Data represent mean ± standard deviation (n = 4) from one of three independent experiments. Student's t test. See also Figure S2, Tables S2 and S3.