Figure 4.
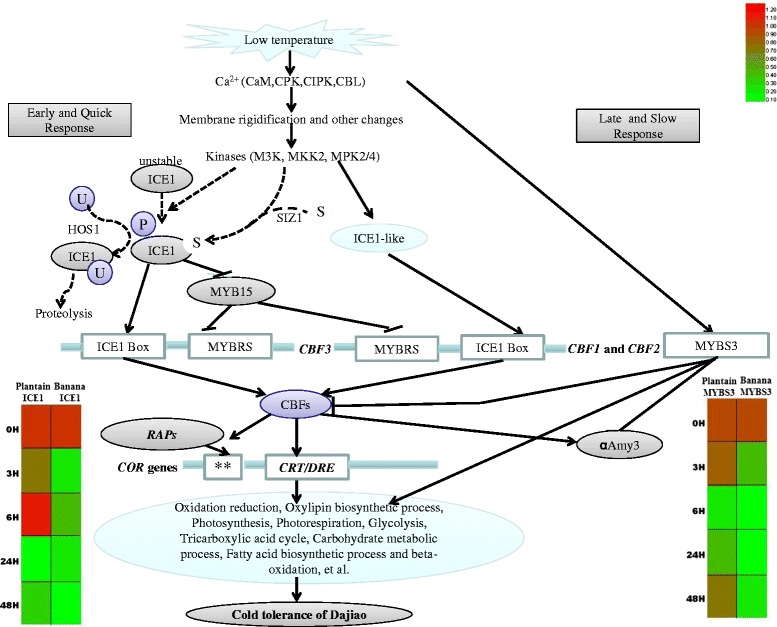
A schematic diagram of cold-tolerance transcriptional network in plantain, adapted initially from V. Chinnusamy et al. (2007) [4] and C.F. Su et al. (2010) [14] and revised based on this study. At the early stage of cold stress, plantain cells probably sense low temperatures through membrane rigidification and/or other cellular changes, which might induce a calcium signature and activate protein kinases necessary for cold tolerance. Constitutively expressed ICE1 is activated by cold stress through sumoylation and phosphorylation. Sumoylation of ICE1 is critical for ICE1-activation of transcription of CBFs and repression of MYB15. CBFs regulate the expression of COR genes that confer cold tolerance. The expression of CBFs is negatively regulated by MYB15. HOS1 mediates the ubiquitination and proteosomal degradation of ICE1 and, thus, negatively regulates CBF regulons. CBFs can constitutively regulate the expression of downstream cold-responsive transcription factor genes RAPs, which might control sub-regulons of the CBF regulon. CBFs also activate αAmy3 expression to hydrolyse reserved starch. At the late stage of cold stress,MYBS3 inhibits CBFs and αAmy3 expression. The effective coordination across the early and late stages of cold stress by at least two different regulatory pathways appears to efficiently regulate the following metabolic pathways including oxidation reduction, oxylipin biosynthetic process, photosynthesis, photorespiration, glycolysis, tricarboxylic acid cycle, carbohydrate metabolic process, fatty acid biosynthetic process and beta-oxidation. The rapid activation and selective induction of ICE1 and MYBS3 cold tolerance pathways in plantain, along with expression of other cold-specific genes, may be one of the main reasons that plantain has higher cold resistance than banana (Heatmaps show the expression of ICE1 and MYBS3 in banana and plantain under cold stress). Broken arrows indicate post-translational regulation; solid arrows indicate activation, whereas lines ending with a bar show negative regulation; the two stars (**) indicate unknown cis-elements. P, phosphorylation; S, SUMO; U, ubiquitin.
