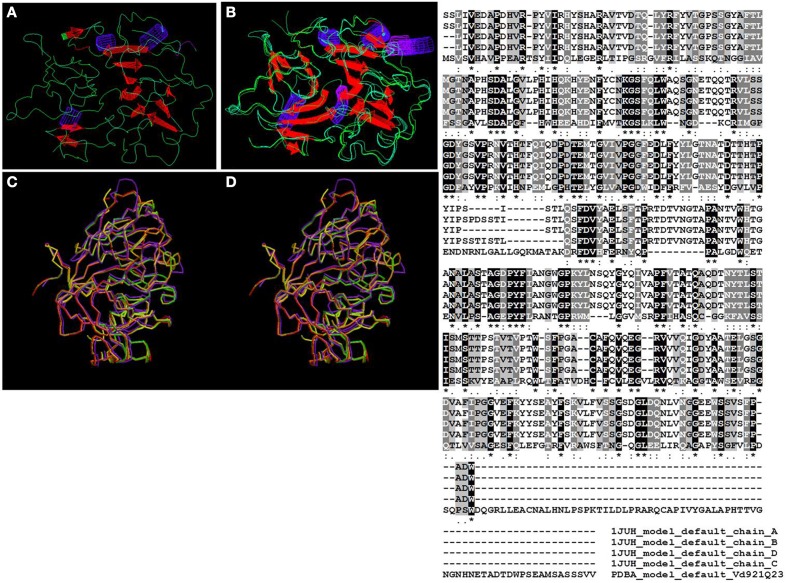
Figure 3.
Theoretical model generated using WhatIf program illustrating the structure of the quercetinase from V. dahliae (A) and its superposition with the template quercetin 2,3-dioxygenase from A. japonicus (B). (C, D) are outputs of the SuperPose program, where (D) represents the V. dahliae protein and (C) the the template quercetin 2,3-dioxygenase from A. japonicus. Minor conformational changes can be noticed between the two structures. The ClustalW2 alignment conducted by the SuperPose program during the superposition of the quercetinase protein from V. dahliae (PDBA_Vd921Q23) and the template quercetin 2,3-dioxygenase from A. japonicus (1JUH_), which have four chains labeled (A–D) highlights the high degree of conservation between the two structures. “*” means that the residues in the column are identical in all aligned sequences; “:” means that conserved substitutions have been observed; “.” means that semi-conserved substitutions are observed; “-” means that there is a gap between the aligned sequences.