Fig 2.
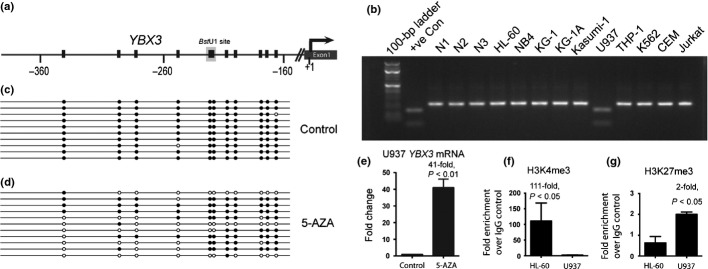
Silencing of YBX3 in U937 cells is associated with promoter DNA hypermethylation and altered histone modifications. (a) YBX3 promoter showing individual CpGs (vertical black line) and their distance from the transcriptional start site (+1). Recognition site for BstU1 used in combined bisulfite restriction analysis (COBRA) assays for YBX3 is shown on the map. (b) Gel electrophoretogram of COBRA showing PCR amplicons of the YBX3 promoter following bisulfite conversion and digestion with BstU1 of genomic DNA from leukemic cell lines and controls. (c) Clonal bisulfite sequencing confirming hypermethylation at the YBX3 promoter in U937 cells. Each row represents a single cloned PCR amplicon aligned to the map shown in (a). Black and white circles denote methylated and unmethylated CpGs, respectively. (d) Reversal of DNA hypermethylation in 5-Aza-2′deoxycytidine (5-AZA)-treated U937 cells as detected by clonal bisulfite sequencing. (e) Quantitative RT-PCR showing increased expression of YBX3 following treatment of U937 cells with 5-AZA. (f) Fold enrichment of YBX3 promoter sequence bound to H3K4me3 (activation mark) and (g) H3K27me3 (silencing mark) in HL-60 and U937 cells normalized to IgG control. +ve Con, positive control, completely methylated human genomic DNA; N1, N2, N3, negative controls, peripheral blood DNA from three healthy individuals.
