ABSTRACT
Microbial symbionts provide benefits that contribute to the ecology and fitness of host plants and animals. Therefore, the evolutionary success of plants and animals fundamentally depends on long-term maintenance of beneficial associations. Most work investigating coevolution and symbiotic maintenance has focused on species-level associations, and studies are lacking that assess the impact of bacterial strain diversity on symbiotic associations within a coevolutionary framework. Here, we demonstrate that fitness in mutualism varies depending on bacterial strain identity, and this is consistent with variation shaping phylogenetic patterns and maintenance through fitness benefits. Through genome sequencing of nine bacterial symbiont strains and cophylogenetic analysis, we demonstrate diversity among Xenorhabdus bovienii bacteria. Further, we identified cocladogenesis between Steinernema feltiae nematode hosts and their corresponding X. bovienii symbiont strains, indicating potential specificity within the association. To test the specificity, we performed laboratory crosses of nematode hosts with native and nonnative symbiont strains, which revealed that combinations with the native bacterial symbiont and closely related strains performed significantly better than those with more divergent symbionts. Through genomic analyses we also defined potential factors contributing to specificity between nematode hosts and bacterial symbionts. These results suggest that strain-level diversity (e.g., subspecies-level differences) in microbial symbionts can drive variation in the success of host-microbe associations, and this suggests that these differences in symbiotic success could contribute to maintenance of the symbiosis over an evolutionary time scale.
IMPORTANCE
Beneficial symbioses between microbes and plant or animal hosts are ubiquitous, and in these associations, microbial symbionts provide key benefits to their hosts. As such, host success is fundamentally dependent on long-term maintenance of beneficial associations. Prolonged association between partners in evolutionary time is expected to result in interactions in which only specific partners can fully support symbiosis. The contribution of bacterial strain diversity on specificity and coevolution in a beneficial symbiosis remains unclear. In this study, we demonstrate that strain-level differences in fitness benefits occur in beneficial host-microbe interactions, and this variation likely shapes phylogenetic patterns and symbiotic maintenance. This highlights that symbiont contributions to host biology can vary significantly based on very-fine-scale differences among members of a microbial species. Further, this work emphasizes the need for greater phylogenetic resolution when considering the causes and consequences of host-microbe interactions.
INTRODUCTION
Mutually beneficial symbiosis (i.e., mutualism) occurs in all domains of life and ecosystems, and its maintenance is critical to plant and animal ecology (1), evolution (2), and health (3–5). Coevolution (reciprocal evolution) and coadaptation (coordinated mutual changes of traits) between partner pairs shape long-term maintenance of mutualisms and lead to specificity between hosts and symbionts, in that only particular potential partners can fulfill the needs of the symbiosis. Significant insights into mutualism have been gained by studying specificity and coevolution at the species level or higher (6–8). However, animal- and plant-associated bacterial strains (members of a bacterial species as defined by molecular methods) can exhibit dramatic genomic and functional differences (9, 10). Also, host genotype-symbiont genotype-environment interactions likely influence coevolution, according to geographic mosaic theory (11). Indeed, within host-microbe associations, host genotype-symbiont genotype differences in mutualistic success have been identified (12–14). Furthermore, experimental studies have demonstrated coevolution between bacterial symbionts and nonnative hosts such that they better engage in symbiosis (15), as well as the influence of coevolution on competition among symbionts (16). However, lacking are studies that experimentally examined the influence of symbiont strain variation on specificity within a coevolutionary context. Such studies are necessary to better understand how symbiont diversity impacts coadaptation, coevolution, and mutualism maintenance. In this study, we assessed the influence of bacterial strain diversity on specificity and coevolution in the association between Xenorhabdus bovienii bacterial symbiont strains and Steinernema spp. nematodes (Nematoda: Panagrolaimomorpha).
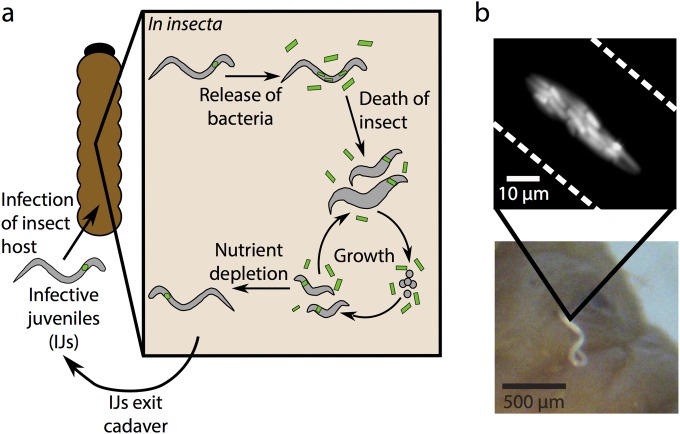
The life cycles of Steinernema spp. nematodes and their proteobacterial symbionts of the genus Xenorhabdus (17) include requisite reproductive stages that occur within an insect host and an environmental nematode stage, the infective juvenile (IJ) (Fig. 1) (18). IJs seek out and invade the insect host, and the nematodes and bacteria kill the insect and reproduce within the cadaver (18, 19). When nutrients are depleted, the nematode progeny form the next generation of IJs (20). Previous studies on a selection of Steinernema-Xenorhabdus pairs have shown that the bacterium can contribute to virulence toward the insect host (21, 22) and support nematode reproduction (19), but it is not known if these bacterial contributions occur with other nematode-bacterium pairs. This interaction is an obligate mutualism in some associations (17), where the nematodes cannot reproduce without the bacterial symbiont (Fig. 1a), and bacteria rely on the nematode host for transmission between insect hosts (Fig. 1b). Xenorhabdus bovienii bacterial strains are broad-host-range symbionts that associate with at least nine Steinernema nematode species from two phylogenetic subclades. Conversely, each of the nematode host species harbors only X. bovienii (23). This suggests that symbiotic maintenance and coevolution may depend on the bacterial subspecies (strain)-level diversity. To assess the impact of bacterial strain diversity within an evolutionary context, we integrated phylogenetic analysis and experimental crossing of symbionts to analyze the coevolutionary relationship and specificity between X. bovienii bacterial strains and their associated nematode hosts.
FIG 1 .
X. bovienii bacterium-Steinernema spp. nematode-Galleria mellonella insect interactions. (a) Schematic diagram of the nematode (gray)-bacterium (green) life cycle. All the events shown within the box occur within the insect host. (b) Image of interactions, provided to show the actual scale. The photograph shows an infective juvenile nematode that is newly emerged from a G. mellonella cadaver. The inset depicts a micrograph of bacteria within the nematode receptacle. Bar, 10 µm.
RESULTS
Genetic diversity of X. bovienii strains.
Genetic variation among possible partner pairs is a prerequisite for specificity in bacterium-host associations. To begin to understand the extent of genetic variation among X. bovienii strains, we conducted genomic comparisons of nine bacterial strains identified as X. bovienii through 16S rRNA sequencing (see Table S1 in the supplemental material), isolated from six different nematode host species (24, 25). We sequenced and annotated the genomes, and the conserved homologs were compared for average nucleotide identity (ANI). All X. bovienii strains had ANIs greater than 96% (see Table S2 in the supplemental material), further supporting that they belong to a single species according to current standard criteria in microbial systematics (26).
To discern conservation and variation in gene content in the nine X. bovienii strains, we conducted gene content analyses using the MaGe platform (27, 28). We found that approximately 55% of the genome content was shared by all strains, and approximately 94% was shared with at least one other strain, with a range of <1% to 9% of genes being unique to an individual strain (see Table S3 in the supplemental material). Since these unique genes may represent specificity determinants, we analyzed their possible functions suggested by their annotation (see Table S3). Within each strain, the most highly represented gene category was that of unknown function. Additionally, unique genes in some strains were predicted to encode membrane or secreted proteins (e.g., putative toxins) that might be expected to interact with nematode or insect hosts (see Table S3). Other genes identified in this analysis included those of predicted metabolic or cellular function and mobile genetic elements (see Table S3). Although examination of the flexible gene content of the X. bovienii strains awaits a more detailed analysis, in total these data indicated a broad spectrum of genetic diversity, and therefore differences in functional potential among X. bovienii bacterial strains. These results strengthen the idea that specificity and coevolution among partner pairs may be occurring between Steinernema spp. hosts and their native X. bovienii partners.
Cophylogenetic analysis.
Specificity between partner pairs is expected to arise through coevolution occurring during long-term maintenance of symbioses. Therefore, to focus our attention on those partner pairs most likely to exhibit specificity, we used phylogenetic analyses to assess the potential for coevolution, as evidenced by cocladegenesis (identical topologies) between symbiont and host phylogenetic trees. A previous phylogenetic study using a multilocus approach revealed instances of codiversification (similar but not identical tree topologies) in some but not all X. bovienii-Steinernema spp. associations, suggesting that fine-scale specificity between hosts and symbionts may occur (24). To generate more refined phylogenetic inferences, we used conserved homologs from whole bacterial genome content to construct the bacterial phylogeny. Cophylogenetic analyses (Fig. 2) revealed congruencies between the X. bovienii phylogeny (see Fig. S1a in the supplemental material) and the Steinernema host phylogeny (see Fig. S1b). Specifically, the cophylogeny provided robust support for cocladogenesis between the group of S. feltiae and S. puntauvense bacterial symbiont strains, which was supported by statistical evidence. For the nine X. bovienii strain genomes, there are 2,027,025 possible rooted trees. Of these, only 14,175 (0.7% of the total) contained a monophyletic clade consisting of the four bacterial strains from the S. feltiae and S. puntauvense nematode isolates. Furthermore, in only 945 of the possible trees (0.05% of the total) did the topology of the bacterial tree match identically with that of the nematode tree, indicating that the bacterial monophyletic clade is unlikely to have occurred by random chance. This statistical permutation test provided strong evidence that the S. feltiae and S. puntauvense nematode isolates and their bacterial symbiont strains share an evolutionary history, and it is likely that specificity is occurring between these partner pairs. In contrast, among other nematode isolates and bacterial strains, host switching has occurred, including one bacterial strain, Xb-Si, which associates with a nematode of another clade, S. intermedium. Host switching within some but not all parts of the nematode-bacteria phylogeny indicates that not all pairings are strictly maintained. Therefore, there are likely divergent maintenance pressures that impact the different nematode-bacterium pairings.
FIG 2 .
Cophylogeny of nematode species hosts and bacterial symbiont strains. Numbers indicate bootstrap values, and thicker lines indicate posterior probabilities greater than 0.9. All posterior probabilities were greater than 0.8. Shaded boxes indicate clades of nematodes with their respective symbionts: X. bovienii nematode hosts (gray), S. feltiae and S. puntauvense nematodes (green), and the distantly related nematode S. intermedium (yellow).
Experimental testing of mutualistic interactions.
As an experimental test of specificity and coevolution, we examined the ability of nematode-bacterium pairs to engage in mutualism through experimental coinjections of three S. feltiae nematode isolates and nine X. bovienii bacterial strains (see Fig. S2 in the supplemental material). Nematodes and bacteria were reared separately, mixed, and coinjected into Galleria mellonella (Lepidoptera: Pyrallidae) larvae. The progression of the life cycle (Fig. 1a; see also Fig. S2) was then monitored for virulence (percent mortality of the coinjected insects), productive infection (percentage of insect cadavers that produced progeny), progeny number (average number of progeny per productive infection), progeny infective potential (ability of the progeny IJs to seek, invade, and kill insect hosts), and bacterial carriage (average CFU per IJ). To assess these traits within a phylogenetic framework, we used linear regression to compare the measurements to bacterium and nematode phylogenetic distances and Bayesian tree distances (i.e., branch lengths from the consensus Bayesian tree between the native pair and the tested bacterial strain or its host). This analysis enabled the comparison of the experimental data to the strongest phylogenetic trend within the bacterial strains to link performance in mutualism with evolutionary history. Certain nematode-bacterium combinations displayed a large amount of trial-to-trial variability, possibly due to seasonal fluctuations or the use of outbred insect and nematode lines. Due to the lack of an inbred insect line, we controlled for differences among insect hosts by limiting insect size variation and randomizing insects among treatments. To address genetic differences between biological replicates and to control for seasonal fluctuations, data were normalized relative to the native combination within each experiment. While variation remained high in some cases, trends (e.g., slopes of the linear regression lines) were similar across trials.
With respect to virulence, some combinations showed significant differences when compared by log-rank analysis, measuring the overall trends within a survival curve (see Table S4 in the supplemental material). However, linear regression analysis did not show significant correlations between the phylogenetic framework and individual measurements of virulence, such as LT50 values (e.g., the time at which half the insects had died), which suggests that the differences do not reflect coevolution (see Table S5 in the supplemental material). As expected, S. feltiae FL and S. feltiae FR nematodes alone killed fewer insects than nematode-bacterium combinations, indicating X. bovienii bacteria contribute to virulence (see Table S4) (P < 0.05). Similarly, S. feltiae MD nematodes were significantly more virulent by log-rank test when associated with six of the bacterial strains (see Table S4) (P < 0.05), though there was no significant difference when data for these nematodes were combined with the three other strains (see Table S4). This suggests that S. feltiae MD is less reliant than the other nematodes on its bacterial symbiont for virulence. By 7 days postinjection, there were no significant differences among the nematode-bacterium combinations in the percentages of insects they killed (see Table S4). Therefore, minor differences in virulence among the nematode-bacterium combinations likely do not significantly impact the mutualism.
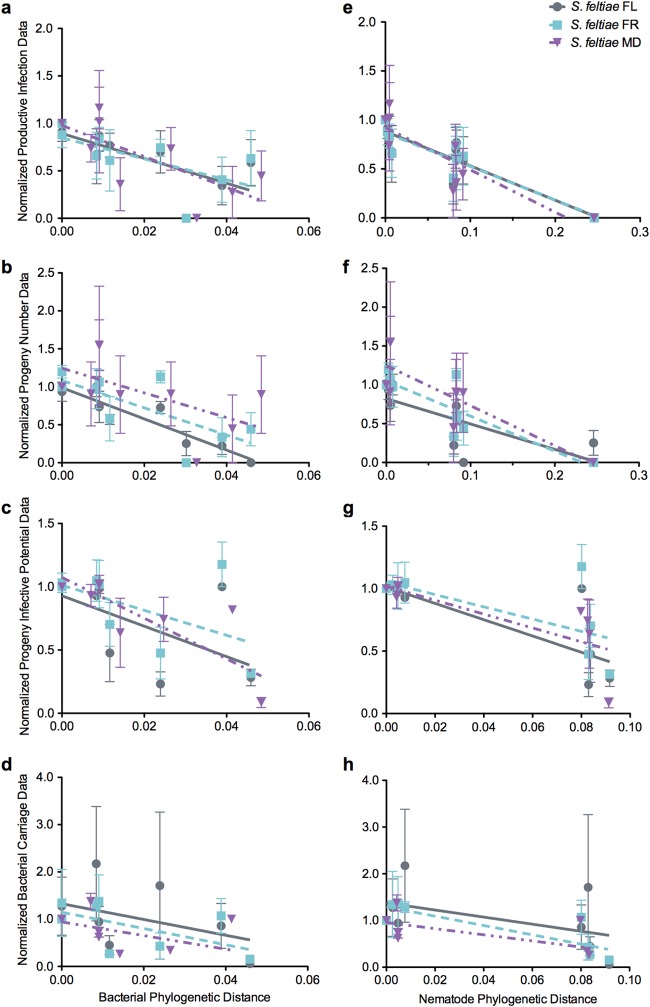
With regard to nematode progeny characteristics, significant differences were identified among different nematode-bacterium pairings relative to bacterial phylogenetic distance or to divergence from the native symbiont. Injections of nematodes alone resulted in no production of progeny, supporting that bacterial symbionts contribute to S. feltiae nematode reproduction (data not shown). Both the ability to produce progeny (productive infection percentage and progeny number) and the infective potentials of those progeny correlated with the bacterial phylogenetic distance; as bacterial strain divergence increased, the success of the interaction decreased (with one exception, the number of S. feltiae MD progeny) (Fig. 3a to c; see also Table S5 in the supplemental material). Although bacterial carriage in nematode progeny also showed a negative trend with respect to phylogenetic distance (Fig. 3d), this was not significant (see Table S5). Productive infection percentage, progeny number, and progeny infective potential were also strongly correlated with nematode host phylogenetic distance (Fig. 3e to g; see also Table S5), indicating that bacteria from hosts more closely related to the S. feltiae nematodes are better able to engage in symbiosis than bacteria from more distantly related hosts.
FIG 3 .
Measurement of mutualistic interaction parameters. Individual parameters of the progression of the nematode-bacterium life cycle were measured with respect to bacterial and nematode phylogenetic distance, including productive infection percentage (a and e), progeny number (b and f), progeny infective potential (c and g), and bacterial carriage (d and h). Phylogenetic distance was taken as the Bayesian tree distance from the native combination data points, representing averages of the measurements for each set of data (n = 3), and error bars represent standard errors. Lines show linear regression for each set of data points (see also Table S5 in the supplemental material). All regressions indicated a significant negative correlation (r2 > 0.2, P < 0.01) for productive infection percentage, progeny number, and progeny infective potential, except for progeny number of S. feltiae MD (r2 = 0.11, P = 0.089). The correlation of bacterial carriage for S. feltiae FR and S. feltiae MD with respect to nematode phylogenetic distance was also significant.
Because the infective potential of the nematode progeny is integral to subsequent nematode reproduction and therefore evolutionary success, we examined potential causes of defects in nematode progeny infective potential. However, we found no significant correlation between bacterial phylogenetic distance and progeny IJ longevity (see Fig. S3a in the supplemental material) or IJ development, as assessed by morphology (see Fig. S3b and c), indicating these parameters do not affect progeny infective potential.
Nematode and bacterial strain fitness.
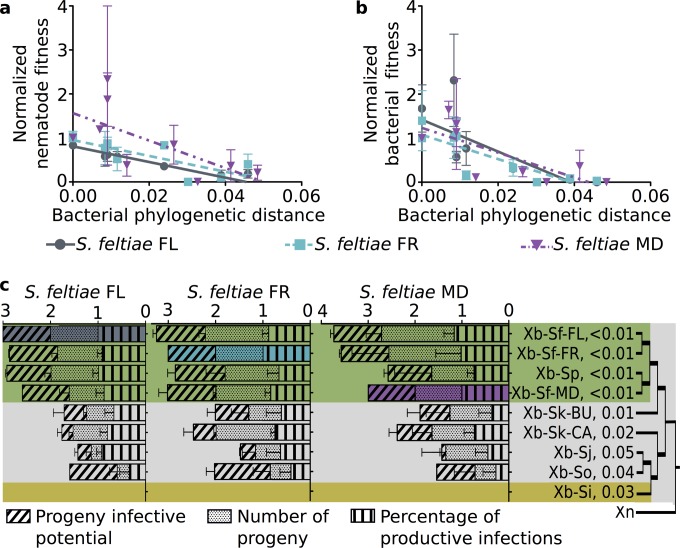
Experimental testing suggests that interactions between the nematode host and bacterial strains closely related to its native symbiont are more efficacious than those with divergent symbionts. We evaluated this trend by using linear regression analysis of nematode reproductive fitness (i.e., total number of progeny, the product of the productive infection percentage and progeny number) relative to bacterial (Fig. 4a; see also Table S5 in the supplemental material) and nematode (see Table S5) phylogenetic distances. There was a significant negative relationship for each nematode isolate (see Table S5), demonstrating that nematode fitness is highest with strains closest to its native bacterial partner or isolated from closely related hosts.
FIG 4 .
Nematode and bacterial combinations show measurements consistent with coadaptation. Graphs show linear regression analysis results of normalized values of nematode (a) and bacterial (b) fitness relative to bacterial phylogenetic distance. Data points represent averages of the measurements for each set of data (n = 3), and error bars represent standard errors. Lines show linear regression for each set of data points (see also Table S5 in the supplemental material). Both linear regressions had a significant negative correlation (r2 > 0.16, P < 0.04). In the stacked bar graph (c), the differing mutualistic interaction parameters are compared to the bacterial phylogeny, with coloring the same as that used in Fig. 2. Filled bars indicate the natural combination, and values next to terminal branches indicate Bayesian tree distances from Xb-Sf symbionts.
To measure bacterial fitness within each combination, we calculated the total number of bacteria that are transmitted to the next generation. Because bacteria are only transmitted through the IJ progeny, this measure is defined as the product of nematode reproductive fitness and bacterial carriage. Using linear regression, a significant negative trend was observed between bacterial fitness and either bacterial phylogenetic distance (Fig. 4b; see also Table S5 in the supplemental material) or nematode phylogenetic distance (see Table S5), indicating that bacterial fitness when in association with S. feltiae is proportional to how similar the bacterium is to the native symbiont and how similar its native host is to S. feltiae. Although absolute bacterial carriage per IJ showed a negative trend observed among strain combinations, the correlation was not significant (Fig. 3d and h; see also Table S5), indicating nematode reproductive fitness contributes more than carriage rate to bacterial fitness.
The data show a clear trend that strains within the same phylogenetic clade as the natural symbiont provide greater benefits than those from different clades (Fig. 4c). This indicates that closely related strains (those that fall within the same clade as the native symbiont) may be functionally redundant in hosts, which in turn suggests that S. feltiae and S. puntauvense nematode isolates require the same goods and services from their symbiont. In contrast, more distantly related strains were defective in various parameters of mutualistic support of nematode fitness (e.g., Xb-Sj has a greater defect in progeny infective potential than Xb-So, which has a greater defect in productive infection percentage), suggesting that not all X. bovienii bacterial strains are functionally redundant within nematode hosts (Fig. 3 and 4c).
The observed negative trends might be caused by bimodal or distributed variations in strain contributions to fitness. Distributed variation is supported by the fact that negative trends remained similar (e.g., negative slope of the same magnitude) when only the symbionts outside the native clade were considered (data not shown). To examine this further, the effects of the native clade data points on the linear negative trends relative to bacterial phylogenetic distance (Fig. 3; see also Table S5 in the supplemental material) were analyzed by computing Cook’s distance (29). Linear regressions for each trend containing all 5 strains outside the native clade and one data point representing the average of all symbionts from the native clade were constructed and used for the analyses. In all trends tested but one, the Cook’s distance of the point representing the native symbiont clade was less than 1, indicating that this data point was not influential on the overall trend (see Table S6 in the supplemental material). Therefore, the observed trends depend on the various results from all bacterial strains, not just a gross difference between bacteria that fall within the clade versus those outside the clade. The one trend that was dependent on inclusion of the native symbiont clade in the analyzed data was bacterial carriage by S. feltiae MD. However, the high variability of the data underlying this particular trend likely caused dramatic impacts on regression curves when individual data points were removed.
X. bovienii bacterial genes contributing to specificity.
Fitness benefit differences among the bacterial strains are unexpected in light of their close phylogenetic relationship; one explanation for the observed fitness advantages of X. bovienii within the S. feltiae and S. puntauvense clade symbionts is variation in shared bacterial genes which could alter functionality and/or cause incompatibility between hosts and symbionts. Such genes are expected to be under positive selection across all bacterial symbionts or within a specific group, such as the S. feltiae and S. puntauvense clade. However, due to the close similarities of the bacterial strains, we were unable to reliably detect positive selection through dN/dS analysis (analysis of the ratio of nonsynonymous to synonymous evolutionary changes).
Another explanation is that S. feltiae and S. puntauvense symbionts as a group uniquely encode or lack genes that impact fitness. The increased fitness of these symbionts relative to other strains may be due to the presence of genes that are absent in other bacterial strains that increase their ability to support symbiotic interactions or the absence of genes that are present in other bacterial strains that have deleterious effects on the nematode host. To assess which genes follow this pattern, we identified genes, based on sequence identity and clustering, that were significantly overrepresented or underrepresented in the symbionts of S. feltiae and S. puntauvense compared to the other X. bovienii strains (see Fig. S4 in the supplemental material). Among the identified families, the most common were of unknown function, representing 80% of overrepresented genes and 54% of underrepresented families. Families of mobile genetic elements were the second most abundant, comprising 4% of overrepresented genes and 38% of underrepresented genes. Other families were those predicted to be involved in primary and secondary metabolism, toxins, and toxin/antitoxin systems. Genes involved in metabolism are potentially important in the symbiosis due to their possible nutritional role. Among the overrepresented metabolic genes (see Fig. S4) were two (azlC and azlD) predicted to encode components of a branched-chain amino acid transporter and one predicted to encode an associated transcriptional regulator (rhaR). Genes from these three families, as well as another of unknown function (listed in the metabolism group), are clustered together in a putative operon that is only present in the S. feltiae and S. puntauvense symbionts. Toxins may also be involved in symbiosis specificity due to their potential role in defensive symbiosis. Additionally, toxin/antitoxin modules have been implicated in ensuring symbiont transmission through regulation of symbiosis genes (30). However, elucidation of the roles of these gene families awaits further functional analyses.
The presence or absence of specific genes within the S. feltiae and S. puntauvense clade symbionts likely reflects the coevolution of these symbionts and their specific hosts. It is possible that the symbionts within the clade have gained or lost certain genes due to the requirements of the nematode host. However, it is also possible that the nematode host has evolved in response to bacterial products (e.g., now the host requires nutrients because the bacterium could produce them). In either case, the partners are a coevolved unit.
DISCUSSION
The observed negative correlation between symbiotic success and the phylogenetic framework supports two overarching ideas. First, the correlation suggests that in the S. feltiae-X. bovienii symbiosis, specificity likely is due to coadaptation between symbionts and hosts, with the bacterial symbiont and/or the host having coevolved to fill the needs of the other partner. Additionally, the fact that the observed fitness trends are inversely correlated with nematode host phylogenetic distance, in some cases more strongly than with bacterial phylogenetic distance (see Table S5 in the supplemental material), provides compelling evidence that the nematode host with which a symbiont associates is a contributing factor to the correlation. This idea is further supported by our finding that the bacterial strain that associates with the most phylogenetically distant nematode host, Xb-Si, is unable to engage in any aspect of mutualism with S. feltiae, even though it is not the most distantly related bacterium from the native symbiont (Fig. 4c). Xb-Sj is the most distantly related symbiont, but it is able to engage in symbiotic interactions (Fig. 3 and 4c).
The second fundamental concept supported by our experimental data is that animal hosts can receive significantly different fitness benefits when associating with divergent bacterial symbiont strains, and such variations could contribute to the evolution of specificity within a symbiotic relationship. Coevolution and coadaptation may be caused by nematode adaptation to unique provisions of particular bacterial strains or by bacterial strain adaptation to fulfilling the specialized needs of a nematode host. In either case, the nematode-bacterium pairings are coevolved units; their life histories are intertwined, and the fitness of each depends on their combined success. The two overarching ideas supported by our data are likely true for other symbioses. Indeed, recent studies have revealed that strain-level symbiont variability impacts the success of the squid-Vibrio (14) and legume-Rhizobium (12, 13) symbioses, although these studies did not assess the impact of variability with relation to coevolution.
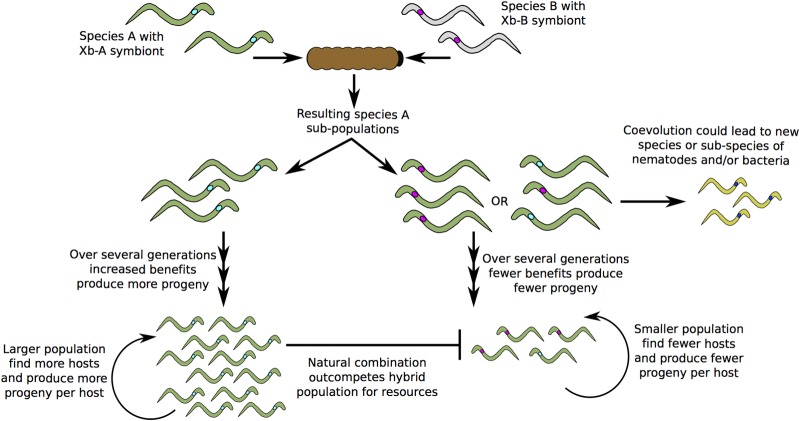
Our data are based on experimental laboratory crosses, but the findings are likely to be ecologically relevant, as mixed infections with multiple nematode hosts and symbionts can occur (31). In fact, nematode isolates of three of the host species relevant to our study (S. feltiae, S. kraussei, and S. intermedium) have been isolated in close proximity (32), although the isolates used here were not. Because mixed infections can lead to nonnative crosses of nematode hosts and bacterial symbionts, variations in benefits could play a significant role in driving maintenance of partner pairs. Greater benefits of some associations relative to others could provide positive feedback for the evolutionary maintenance of the mutualism (i.e., increased evolutionary success) and therefore a selective pressure that results in the observed phylogenetic patterns (Fig. 5). This is consistent with the theory of partner fidelity feedback, where linkages in host and symbiont fitness provide positive reinforcement that stabilizes the mutualism (2). A partner fidelity feedback mechanism is further supported by the fact that S. feltiae nematodes and X. bovienii bacteria cannot separately complete their life cycles (Fig. 1b and data not shown). Our data extend current theory by demonstrating that bacterial strain diversity can drive partner fidelity feedback for maintenance of mutual relationships.
FIG 5 .
Contributions of increased benefits in maintenance of symbiosis. The illustration shows how increased benefits of the native pairing could serve to reinforce partner fidelity within a symbiotic relationship and therefore contribute to symbiotic maintenance. The different colors represent divergent nematode species or bacterial strains.
Diversity in the goods and services that are integral to symbiotic function are likely responsible for variations in symbiotic success. One mutualistic role that bacterial symbionts can perform is defense, where the symbiont provides protection for the host against specific pathogens, competitors, or predators. For example, this type of function can be conferred by secondary symbionts of aphids, where the bacterial symbiont contributes to protection against a parasitoid wasp predator of the aphid host. In some of these associations, protection is conferred by a phage-carried toxin, but this mechanism is not conserved among all secondary symbionts (33). Similarly, specialized toxins or secondary metabolites produced by some but not all X. bovienii may contribute to nematode host defense against organisms within the insect host, as is true for X. nematophila (34, 35). In the absence of the defensive compound, the nematode host may be killed before establishing a new infection or suffer from population losses that result in fewer progeny. Alternatively, since some classes of Xenorhabdus toxins are insecticidal (36), variations in symbiont-encoded toxins could influence the efficiency of insect host killing, as well as the insect host range of the nematode. Finally, symbiont-derived toxins may have nematicidal effects to which the native nematode host is resistant. In a nonnative pairing, such a nematicidal activity obviously would have a detrimental impact.
Many of the genes identified as unique (see Table S3 in the supplemental material) or enriched (see Fig. S4 in the supplemental material) within the X. bovienii bacterial strains could encode defensive factors. For example, unique and enriched genes including those encoding exported proteins of unknown function, nonribosomal peptide synthetases (NRPS), polyketide synthetases (PKS), and a predicted Shiga toxin. NRPS-PKS gene clusters are of particular interest, as they synthesize small molecules (secondary metabolites) that have a variety of biological activities, such as antimicrobial (37–39), hemolytic (39), and immunosuppressive (40) effects, which could serve as defensive factors. Indeed, a fungal endosymbiont NRPS gene cluster provides plants protection from insect feeding (41). Also of interest is the putative Shiga toxin that, among the tested X. bovienii strains, is uniquely encoded by Xb-Si. Shiga toxins affect lipid bilayers and have cytotoxic activities that allow bacterial pathogens to cause disease within eukaryotic hosts (42). Such a toxin could be involved in pathogenesis toward the insect host or in defense. Indeed, the presence of this toxin may be responsible for the inability of Xb-Si to support symbiosis with S. feltiae nematodes (Fig. 4c), as it could have a cytotoxic effect in the S. feltiae nematodes.
Another basis for many symbiotic relationships is the mutually beneficial exchange of nutrients between host and symbiont, and integration of nutritional requirements could influence coadaptation. It has been proposed that Xenorhabdus bacteria provide nutritional support to the nematode host, and lack of this support leads to a decreased ability to produce progeny (19, 43). Consistent with this, S. feltiae nematodes are unable to reproduce within the insect host without their bacterial symbionts (data not shown), possibly due to a lack of specific nutrients. Further corroborating this idea, we observed that among the S. feltiae/S. puntauvense symbionts there was an overrepresentation of genes predicted to encode branched-chain amino acid transporter systems, which could contribute to nutritional support of nematode reproduction or infective potential. Branched-chain amino acid transport influences Rhizobium nodulation on plants, with various phenotypes depending on the plant host (44). Thus, variability in symbiont amino acid transport capability, coupled with variation in nematode host requirements for certain amino acids, could help explain some fitness differences.
There is increasing recognition that beneficial microbes have profoundly impacted the origin and evolution of animals (4), and studies have implicated symbiosis in host speciation through reproductive isolation (8). These phenomena depend on symbioses being maintained throughout the evolutionary history of the host organism. We found that this maintenance is shaped by bacterial strain diversity, and our analyses have highlighted that the contributions of symbionts to host biology can vary significantly based on differences among members of a microbial species. Furthermore, our findings suggest that in addition to current studies of microbiota that identify microbes by the species or phylum level, a comprehensive understanding of the causes and consequences of host-microbe interactions will require the use of greater phylogenetic resolution and classification through evolutionary and functional characterizations.
MATERIALS AND METHODS
Bacterial strains and nematode isolates.
Bacterial strains were obtained by sonication of nematode hosts after surface sterilization. Strain identity was confirmed by analysis of 16S rRNA from whole-genome sequences (25, 43) and average nucleotide identity (26). Bacterial strains were stored in lysogeny broth (LB) supplemented with 20% glycerol frozen at −80°C. Bacterial cultures were grown at 30°C in LB that had not been exposed to light, with aeration, or on LB agar with 0.1% pyruvate (45). S. feltiae nematode isolates used were obtained from the laboratories of Patricia Stock or Byron Adams (via Adler Dillman) and were verified through sequencing of the 12S and 28S genes (24). Nematodes were propagated through Galleria mellonella larvae (46). Axenic IJs were produced in vitro as previously described (47, 48). Nematodes were stored at 25°C in 250-ml tissue culture flasks (BD Falcon, Franklin Lakes, NJ) at a density of 5 to 10 IJ/µl and a volume of less than 60 ml.
Genes and genome sequencing and assembly.
Nematode 12S and 28S rRNA genes were sequenced as previously described (49), and sequences were submitted to NCBI. Bacterial genomes were sequenced using Illumina paired-end libraries (mean insert length, 300 bp) and quality trimmed using DynamicTrim.pl v.1.10 in the solexQC package. Dynamic trimming was performed according to TrimAI’s, v1.2rev59 (50) default settings. After excluding reads of <20 bp and lacking barcode sequences, genomes were assembled with Velvet v.1.1.06 (51, 52) using automatic determination of sequencing coverage and a series of kmer values. Draft genomes were annotated using MaGe (27, 28) and submitted to EMBL. 16S rRNA sequences from draft genomes were assessed using BLASTn (http://blast.ncbi.nlm.nih.gov) and were submitted to GenBank.
Phylogenetic analysis.
For bacterial phylogenetic analysis, MaGe was used to identify homologs (genes with conserved synteny with ≥30% nucleotide identity over ≥80% of the length) present in the X. bovienii and X. nematophila genomes, yielding 2,166 gene sets. Sets were excluded if any genome possessed multiple homologs (putative paralogs), resulting in 1,893 ortholog sets. Nematode genes and ortholog sets were corrected using ORFcor v1.02 (53). The corrected sets were aligned using Muscle v3.7 (54, 55), and poorly aligned regions were excluded using TrimAI v1.2rev59 (50). Genes were concatenated into a single alignment (1,867,725 bp total). Maximum likelihood analyses were performed in RAxML v7.2.8 (56) using the GTR+γ substitution model with rapid bootstrapping of 1,000 replicates, optimized via use of jModelTest v2.1.4 (57). Bayesian analyses were done using MrBayes v3.2.1 (58). For the nematode tree, posterior probabilities were sampled from 4,000,000 MCMC replicates, with the first 1,000,000 discarded as burn-in. Using the GTR+γ substitution model, the final standard deviation of split frequencies was 0.0081. For the bacterial tree, posterior probabilities were sampled from 1,000,000 MCMC replicates, with the first 250,000 discarded as burn-in. The GTR+γ substitution model was used, as were the following nonstandard parameters to optimize MCMC sampling: 2 runs, each with 5 heated and 1 cold chain, Multiplier(V) $lambda = 0.05, and TLMultiplier(V) $lambda = 0.05. Topologies of nematode and bacterial phylogenies were identical from both methods. Distance trees were viewed and drawn by using iTol (59).
Testing of mutualistic interactions. (i) Virulence assays.
Fifth-instar G. mellonella larvae (Grubco, Hamilton, OH) were used for injections. Overnight cultures of bacteria were subcultured 1:100 into LB, grown to an optical density at 600 nm (OD600) of ~0.65, and diluted in Grace’s insect medium (Sigma, St. Louis, MO) to achieve 200 CFU in a 10-µl injection volume. Prior to injections, test OD measurements and CFU counting by dilution plating were done to ensure that equal CFU counts were used for all strains. Axenic nematodes were surface sterilized and diluted in Grace’s medium to inject 50 IJs in 10 µl. For coinjections, bacteria and nematodes were prepared as described above and combined for injection with 200 CFU of bacteria and 50 IJs in 10 µl. Virulence was measured as the percent survival, with assessment every 24 h for 7 days. Insects were considered dead when they no longer responded to gentle prodding. Twelve insects were injected per treatment, and three experimental replicates were performed.
(ii) Productive infection experiments.
Insect cadavers obtained from injection assays were placed individually in a modified White trap (60) at day 7 postinjection and observed for progeny emergence daily for up to 1 month. Cadavers producing at least 100 IJs were scored as productive, and the productive infection percentage was calculated as the number of cadavers producing progeny out of the total number. Three experimental replicates were performed.
(iii) Progeny counts.
The number of IJs produced from each insect cadaver was determined by removing and measuring water from each White trap and counting the number of IJs/ml. IJs were counted every other day for 16 days and on day 28, and the total number of IJs was calculated. Three biological replicates were performed. IJ progeny were collected for 10 days from the average first day of emergence, pooled, and stored for use.
(iv) Progeny infective potential.
Progeny infective potential was determined through a modified sand trap assay (61), using 50 IJs in 100 µl of water. Two technical replicates of 12 G. mellonella larvae were used per pool of progeny. Mortality was monitored every 8 h for 96 h. Three biological replicates were performed. For linear regression, the percent mortality at 48 h was used, as it showed the largest difference between combinations.
(v) Bacterial carriage.
Bacterial symbiont carriage was determined similarly to a previously described method (62) through surface sterilization, grinding of progeny nematodes, and dilution plating. Two technical replicates and three biological replicates were performed.
(vi) Progeny IJ longevity.
Progeny IJs were stored for 6 months postemergence and then examined at 3× magnification. Dead nematodes were straight and/or showed no movement, whereas live nematodes had a curved appearance or were moving. IJs were confirmed as dead by gentle prodding. Two technical replicates of at least 100 IJs were counted, and three biological replicates were performed.
(vii) Morphometric analysis.
Ten-milliliter aliquots of progeny IJs were heat killed in M9 buffer at 60°C. Heat-killed IJs were fixed in triethanolamine formalin (TAF) at 60°C (63) and mounted on glass slides for morphometric analysis. Quantitative measurements (length and width) were made using an Olympus BX51 microscope with Olympus Microsuite software (Soft Imaging System Corp., CA, USA).
Comparative genomics. (i) dN/dS analysis.
dN/dS analysis was done using PAML v4.7 (64). Protein sequences of bacterial homologs (see “Phylogenetic analysis,” above) were aligned using the e-nsi-I algorithm implemented in MAFFT v7.029 (65) and converted to codon alignments. The branch model was used to test for positive selection in the X. bovienii clade, and a dN/dS cutoff of 1 to 6 with at least 2% amino acid divergence was used (66).
(ii) Overrepresented and underrepresented gene sets.
Protein sequences were downloaded from MaGe and annotated using HMMer models for KEGG (http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=24214961) and Pfam (67). Sequences were clustered de novo, based on amino acid sequence identity (25% sequence identity and 50% coverage), using the proteinorthov5 program available via the BioMed Central website. Fisher’s exact test, implemented within the python package scipy (http://www.scipy.org), was used to identify gene families or domains that were over- or underrepresented in a particular clade. A cutoff of P < 0.05 was used to determine significance.
Statistics.
Cophylogeny was statistically measured by a permutation test. The total number of possible X. bovienii trees was compared to the number of possible trees that contained the monophyletic group of S. feltiae and S. puntauvense symbionts and to the number of possible trees with identical topology in the bacterial monophyletic group (68). The number of trees with a given split was the product of the number of rooted trees for each half.
Statistical analyses for experimental testing were performed using R (69) or Prism (GraphPad). For virulence assays, a log rank test and LT50 calculation were done. In the log rank analysis, all data points from the three experimental replicates were combined. Each experimental block was also analyzed separately, and trends remained the same. For experimental testing of mutualistic interactions, linear regression analyses were done to determine R2 values and the P values for trends. The data were normalized to the native combination (all values were divided by the value of the natural combination). Technical replicates were averaged and plotted versus the phylogenetic distance of the bacterial symbiont or nematode host of the bacterial symbiont, the Bayesian tree distance from the native combination. The data were also compared to ANI and maximum likelihood tree distances with similar results. Cook’s distance for each trend was computed using a linear regression constructed based on all symbionts outside the native clade (i.e., Xb-Si, Xb-Sj, Xb-Sk-BU, Xb-Sk-CA, and Xb-So) plus one data point of the average of the native clade (i.e., Xb-Sf-FL, Xb-Sf-FR, Xb-Sf-MD, and Xb-Sp). A Cook’s distance of >1 was considered influential.
Nucleotide sequence accession numbers.
Draft nematode and bacterial genome sequences were annotated and submitted to EMBL, and 16S rRNA sequences from draft genomes were assessed using BLASTn. The sequences were submitted to GenBank; the corresponding accession numbers are provided in Table S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Bayesian distance phylogenies. Bacterial (a) and nematode (b) distance trees are shown. The trees were constructed using Bayesian methods and had the same topology and similar distances as those constructed using maximum likelihood analysis. Download
Schematic diagram of mutualistic testing. The diagram depicts the progression of experimental testing. Axenic IJs (nematodes not exposed to bacteria) and the bacterial symbionts were reared separately, and each combination was mixed and coinjected into the insect host. Insects were monitored for percent mortality (virulence of combination) and progeny nematode IJ emergence (productive infections and progeny number). Progeny IJs were stored in flasks and then used for progeny measurements: progeny infective potential, bacterial carriage, morphometric analysis, and longevity. Download
Possible causes of progeny infective fitness defects were analyzed using linear regression relative to bacterial phylogenetic distance (Bayesian tree distance) for nematode progeny longevity (a) or morphometric analyses (b and c). For morphometric analyses, both length (b) and width (c) were taken into consideration as measurements of nematode progeny health. No significant correlations were found with adequate r2 values. Data points represent averages of the measurements for each set of data (n = 3), and error bars represent standard errors. Download
Heat map of overrepresented and underrepresented genes. To identify nonconserved gene sets potentially involved in the nematode-bacterium mutualism, genes from all X. bovienii genomes were analyzed for overrepresentation (a) or underrepresentation (b) within the monophyletic clade of S. feltiae and S. puntauvense bacterial symbiont strains. The total gene number per symbiont genome is shown in the heat map and is relative to the phylogeny of bacterial symbionts. Each gene family is listed in a separate row, and the predicted function is shown to the left of each row of counts. Download
Bacterial strains and nematode host species used in this study.
Average nucleotide identities of X. bovienii strains used in the experiments.
Unique genomic content differences between X. bovienii strains.
Virulence of S. feltiae nematodes alone and of X. bovienii coinjections.
Linear regression analyses of data from Table S4 and Fig. 3 and 4 relative to bacterial phylogenetic distances.
Cook’s distance for the native symbiont clade average point (derived from linear regression analyses of parameters from Table S5 and Fig. 3).
ACKNOWLEDGMENTS
We thank Adler Dillman, John Chaston, Michael Gebre, and Byron Adams (Brigham Young University) for the S. feltiae MD nematode isolate and Lorena Uribe-Lorío (ICBCM, Universidad de Costa Rica) for the S. puntauvense nematode isolate. We also acknowledge the College of Agriculture and Life Sciences Statistical Consulting Group for support in statistical analyses and MaGe for genome annotation and analysis.
This study was supported by a collaborative grant from the National Science Foundation to H.G.-B. (IOS-0920631), S.F. (IOS-0919912), and S.P.S. (IOS-0919565). K.E.M. was also supported by the National Institutes of Health (NIH) National Research Service award T32 AI55397 and a Louis and Elsa Thomsen Distinguished Predoctoral Fellowship. B.R.M. was supported by NIH National Research Service Award T32 GM07215 and the Department of Energy Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494). J.L.K. was supported by a National Science and Engineering Research Council of Canada (NSERC Canada) postdoctoral fellowship.
Footnotes
Citation Murfin KE, Lee M, Klassen JL, McDonald BR, Larget B, Forst S, Stock SP, Currie CR, Goodrich-Blair H. 2015. Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio 6(3):e00076-15. doi:10.1128/mBio.00076-15.
REFERENCES
- 1.Saavedra S, Stouffer DB, Uzzi B, Bascompte J. 2011. Strong contributors to network persistence are the most vulnerable to extinction. Nature 478:233–235. doi: 10.1038/nature10433. [DOI] [PubMed] [Google Scholar]
- 2.Weyl EG, Frederickson ME, Yu DW, Pierce NE. 2010. Economic contract theory tests models of mutualism. Proc Natl Acad Sci U S A 107:15712–15716. doi: 10.1073/pnas.1005294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg G. 2009. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 4.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 6.Tianero MD, Kwan JC, Wyche TP, Presson AP, Koch M, Barrows LR, Bugni TS, Schmidt EW. 2015. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J 9:615–628 doi: 10.1038/ismej.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brucker RM, Bordenstein SR. 2013. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341:667–669. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 9.Galardini M, Mengoni A, Brilli M, Pini F, Fioravanti A, Lucas S, Lapidus A, Cheng JF, Goodwin L, Pitluck S, Land M, Hauser L, Woyke T, Mikhailova N, Ivanova N, Daligault H, Bruce D, Detter C, Tapia R, Han C, Teshima H, Mocali S, Bazzicalupo M, Biondi EG. 2011. Exploring the symbiotic pangenome of the nitrogen-fixing bacterium Sinorhizobium meliloti. BMC Genomics 12:235. doi: 10.1186/1471-2164-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson J. 2005. The geographic mosaic of coevolution. University of Chicago Press, Chicago, IL. [Google Scholar]
- 12.Heath KD. 2010. Intergenomic epistasis and coevolutionary constraint in plants and rhizobia. Evolution 64:1446–1458. doi: 10.1111/j.1558-5646.2009.00913.x. [DOI] [PubMed] [Google Scholar]
- 13.Thrall PH, Laine AL, Broadhurst LM, Bagnall DJ, Brockwell J. 2011. Symbiotic effectiveness of rhizobial mutualists varies in interactions with native Australian legume genera. PLoS One 6:e23545. doi: 10.1371/journal.pone.0023545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez-Dozal AA, Gorman C, Lostroh CP, Nishiguchi MK. 2014. Gene-swapping mediates host specificity among symbiotic bacteria in a beneficial symbiosis. PLoS One 9:e101691. doi: 10.1371/journal.pone.0101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soto W, Punke EB, Nishiguchi MK. 2012. Evolutionary perspectives in a mutualism of sepiolid squid and bioluminescent bacteria: combined usage of microbial experimental evolution and temporal population genetics. Evolution 66:1308–1321. doi: 10.1111/j.1558-5646.2011.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiguchi MK, Ruby EG, McFall-Ngai MJ. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in Sepiolid squid-Vibrio symbioses. Appl Environ Microbiol 64:3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poinar GO, Thomas GM. 1966. Significance of Achromobacter nematophilus Poinar and Thomas (Achromobacteraceae: Eubacteriales) in the development of the nematode, DD-136 (Neoaplectana sp. Steinernematidae). Parasitology 56:385–390. [DOI] [PubMed] [Google Scholar]
- 18.Herbert EE, Goodrich-Blair H. 2007. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol 5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- 19.Richards GR, Goodrich-Blair H. 2010. Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpA in nematode progeny production. Appl Environ Microbiol 76:221–229. doi: 10.1128/AEM.01715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popiel I, Grove DL, Friedman MJ. 1989. Infective juvenile formation in the insect parasitic nematode Steinernema feltiae. Parasitology 99:77–81. doi: 10.1017/S0031182000061047. [DOI] [Google Scholar]
- 21.Sugar DR, Murfin KE, Chaston JM, Andersen AW, Richards GR, deLéon L, Baum JA, Clinton WP, Forst S, Goldman BS, Krasomil-Osterfeld KC, Slater S, Stock SP, Goodrich-Blair H. 2012. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol 14:924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orchard SS, Goodrich-Blair H. 2004. Identification and functional characterization of a Xenorhabdus nematophila oligopeptide permease. Appl Environ Microbiol 70:5621–5627. doi: 10.1128/AEM.70.9.5621-5627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock SP, Goodrich-Blair H. 2008. Entomopathogenic nematodes and their bacterial symbionts: The inside out of a mutualistic association. Symbiosis 46:65–75. [Google Scholar]
- 24.Lee MM, Stock SP. 2010. A multilocus approach to assessing co-evolutionary relationships between Steinernema spp. (Nematoda: Steinernematidae) and their bacterial symbionts Xenorhabdus spp. (gamma-proteobacteria: Enterobacteriaceae). Syst Parasitol 77:1–12. doi: 10.1007/s11230-010-9256-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee MM, Stock SP. 2010. A multigene approach for assessing evolutionary relationships of Xenorhabdus spp. (gamma-proteobacteria), the bacterial symbionts of entomopathogenic Steinernema nematodes. J Invertebr Pathol 104:67–74. doi: 10.1016/j.jip.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Médigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Médigue C. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook RD. 1977. Detection of influential observation in linear regression. Technometrics 19:15–18. doi: 10.2307/1268249. [DOI] [Google Scholar]
- 30.Nor I, Engelstädter J, Duron O, Reuter M, Sagot MF, Charlat S. 2013. On the genetic architecture of cytoplasmic incompatibility: inference from phenotypic data. Am Nat 182:E15–E24. doi: 10.1086/670612. [DOI] [PubMed] [Google Scholar]
- 31.Koppenhöfer AM, Kaya HK. 1996. Coexistence of two steinernematid nematode species (Rhabditida:Steinernematidae) in the presence of two host species. Appl Soil Ecol 4:221–230. doi: 10.1016/S0929-1393(96)00121-7. [DOI] [Google Scholar]
- 32.Valadas V, Mracek Z, Oliveira S, Mota M. 2011. Three species of entomopathogenic nematodes of the family Steinernematidae (Nematoda: Rhabditida) new to continental Portugal. Nematol Mediterr 39:169–178. [Google Scholar]
- 33.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh S, Reese JM, Casanova-Torres AM, Goodrich-Blair H, Forst S. 2014. Microbial population dynamics in the hemolymph of Manduca sexta infected with Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl Environ Microbiol 80:4277–4285. doi: 10.1128/AEM.00768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales-Soto N, Forst SA. 2011. The xnp1 P2-like tail synthesis gene cluster encodes xenorhabdicin and is required for interspecies competition. J Bacteriol 193:3624–3632. doi: 10.1128/JB.00092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JM, Kim M, Min J, Lee SM, Shin KS, Oh SD, Oh SJ, Kim YH. 2012. Proteomic identification of a novel toxin protein (Txp40) from Xenorhabdus nematophila and its insecticidal activity against larvae of Plutella xylostella. J Agric Food Chem 60:4053–4059. doi: 10.1021/jf204351f. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Gonzalez E, Müller S, Ensle P, Süssmuth RD, Genersch E. 2014. Elucidation of sevadicin, a novel non-ribosomal peptide secondary metabolite produced by the honey bee pathogenic bacterium Paenibacillus larvae. Environ Microbiol 16:1297–1309. doi: 10.1111/1462-2920.12417. [DOI] [PubMed] [Google Scholar]
- 38.Cochrane SA, Vederas JC. 28 May 2014. Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med Res Rev doi: 10.1002/med.21321. [DOI] [PubMed] [Google Scholar]
- 39.Luo C, Liu X, Zhou H, Wang X, Chen Z. 2015. Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl Environ Microbiol 81:422–431. doi: 10.1128/AEM.02921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwecke T, Aparicio JF, Molnár I, König A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortés J, Lester JB. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci U S A 92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. 2005. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol Microbiol 57:1036–1050. doi: 10.1111/j.1365-2958.2005.04747.x. [DOI] [PubMed] [Google Scholar]
- 42.Obrig TG, Karpman D. 2012. Shiga toxin pathogenesis: kidney complications and renal failure. Curr Top Microbiol Immunol 357:105–136. doi: 10.1007/82_2011_172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE, Cowles KN, Darby C, de Léon L, Drace K, Du Z, Givaudan A, Herbert Tran EE, Jewell KA, Knack JJ, Krasomil-Osterfeld KC, Kukor R, Lanois A, Latreille P, Leimgruber NK, Lipke CM, Liu R, Lu X, Martens EC, Marri PR, Medigue C, Menard ML, Miller NM, Morales-Soto N, Norton S, Ogier JC, Orchard SS, Park D, Park Y, Qurollo BA, Sugar DR, Richards GR, Rouy Z, Slominski B, Slominski K, Snyder H, Tjaden BC, van der Hoeven R, Welch RD, Wheeler C, Xiang B, Barbazuk B. 2011. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prell J, Bourdès A, Kumar S, Lodwig E, Hosie A, Kinghorn S, White J, Poole P. 2010. Role of symbiotic auxotrophy in the rhizobium-legume symbioses. PLoS One 5:e13933. doi: 10.1371/journal.pone.0013933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Hurlbert RE. 1990. Toxicity of irradiated Media for Xenorhabdus spp. Appl Environ Microbiol 56:815–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martens EC, Heungens K, Goodrich-Blair H. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol 185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens EC, Goodrich-Blair H. 2005. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell Microbiol 7:1723–1735. doi: 10.1111/j.1462-5822.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- 48.Sicard M, Le Brun N, Pages S, Godelle B, Boemare N, Moulia C. 2003. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: contrasting types of interaction. Parasitol Res 91:520–524. doi: 10.1007/s00436-003-0998-z. [DOI] [PubMed] [Google Scholar]
- 49.Nadler SA, Bolotin E, Stock SP. 2006. Phylogenetic relationships of Steinernema (Cephalobina, Steinernematidae) based on nuclear, mitochondrial, and morphological data. Syst Parasitol 64:159–179. doi: 10.1007/s11230-005-9009-3. [DOI] [PubMed] [Google Scholar]
- 50.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. BioInformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerbino DR. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics Chapter 11:Unit 11.5. doi: 10.1002/0471250953.bi1105s31 PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klassen JL, Currie CR. 2013. ORFcor: identifying and accommodating ORF prediction inconsistencies for phylogenetic analysis. PLoS One 8:e58387. doi: 10.1371/journal.pone.0058387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rokas A. 2011. Phylogenetic analysis of protein sequence data using the randomized axelerated maximum likelihood (RAXML) program. Curr Protoc Mol Biol Chapter 19:Unit19.11. doi: 10.1002/0471142727.mb1911s96. [DOI] [PubMed] [Google Scholar]
- 57.Posada D. 2008. JModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 58.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaya HK, Stock SP. 1997. Techniques in insect nematology. Academic Press, London, England. [Google Scholar]
- 61.Grewal PS, Selvan S, Gaugler R. 1994. Thermal adaptation of entomopathogenic nematodes: niche breadth for infection, establishment, and reproduction. J Therm Biol 19:245–253. doi: 10.1016/0306-4565(94)90047-7. [DOI] [Google Scholar]
- 62.Goetsch M, Owen H, Goldman B, Forst S. 2006. Analysis of the PixA inclusion body protein of Xenorhabdus nematophila. J Bacteriol 188:2706–2710. doi: 10.1128/JB.188.7.2706-2710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Courtney W, Polley D, Miller V. 1955. TAF, an improved fixative in nematode techniques. Plant Dis Rep 39:570–571. [Google Scholar]
- 64.Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 65.Katoh K, Standley DM. 2014. MAFFT: iterative refinement and additional methods. Methods Mol Biol 1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 66.Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol 239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 67.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Felsenstein J. 1978. The number of evolutionary trees. Syst Zool 27:27–33. doi: 10.2307/2412810. [DOI] [Google Scholar]
- 69.R Core Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian distance phylogenies. Bacterial (a) and nematode (b) distance trees are shown. The trees were constructed using Bayesian methods and had the same topology and similar distances as those constructed using maximum likelihood analysis. Download
Schematic diagram of mutualistic testing. The diagram depicts the progression of experimental testing. Axenic IJs (nematodes not exposed to bacteria) and the bacterial symbionts were reared separately, and each combination was mixed and coinjected into the insect host. Insects were monitored for percent mortality (virulence of combination) and progeny nematode IJ emergence (productive infections and progeny number). Progeny IJs were stored in flasks and then used for progeny measurements: progeny infective potential, bacterial carriage, morphometric analysis, and longevity. Download
Possible causes of progeny infective fitness defects were analyzed using linear regression relative to bacterial phylogenetic distance (Bayesian tree distance) for nematode progeny longevity (a) or morphometric analyses (b and c). For morphometric analyses, both length (b) and width (c) were taken into consideration as measurements of nematode progeny health. No significant correlations were found with adequate r2 values. Data points represent averages of the measurements for each set of data (n = 3), and error bars represent standard errors. Download
Heat map of overrepresented and underrepresented genes. To identify nonconserved gene sets potentially involved in the nematode-bacterium mutualism, genes from all X. bovienii genomes were analyzed for overrepresentation (a) or underrepresentation (b) within the monophyletic clade of S. feltiae and S. puntauvense bacterial symbiont strains. The total gene number per symbiont genome is shown in the heat map and is relative to the phylogeny of bacterial symbionts. Each gene family is listed in a separate row, and the predicted function is shown to the left of each row of counts. Download
Bacterial strains and nematode host species used in this study.
Average nucleotide identities of X. bovienii strains used in the experiments.
Unique genomic content differences between X. bovienii strains.
Virulence of S. feltiae nematodes alone and of X. bovienii coinjections.
Linear regression analyses of data from Table S4 and Fig. 3 and 4 relative to bacterial phylogenetic distances.
Cook’s distance for the native symbiont clade average point (derived from linear regression analyses of parameters from Table S5 and Fig. 3).