Figure 1. The mir-35 family is required for embryonic hypoxic survival.
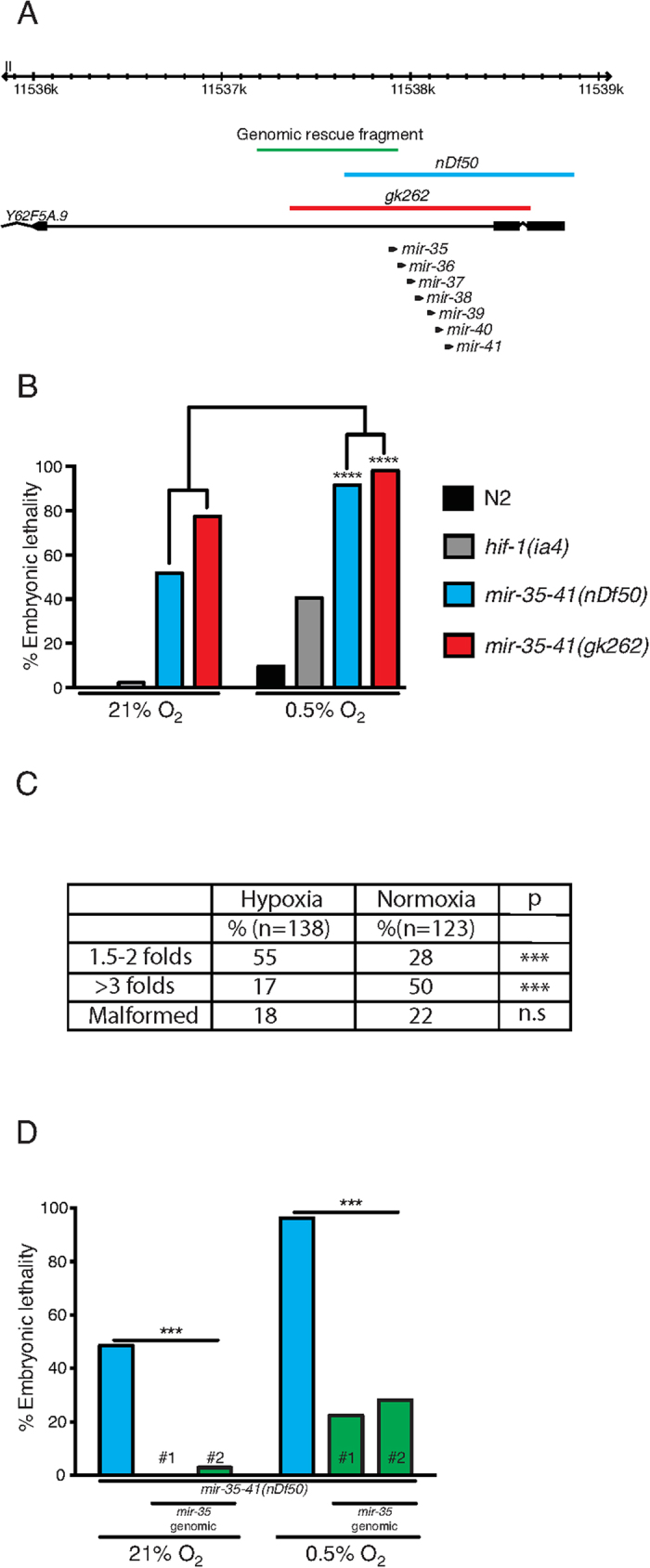
(A) mir-35–41 locus, mutant alleles and genomic rescue fragment. nDf50 (blue) and gk262 (red) alleles remove the entire mir-35–41 locus and part of the Y62F5A.9 gene. The genomic rescue fragment used in (D) is marked in green, which includes a 602 bp upstream region and the mir-35 hairpin. (B) At 21% O2, two mir-35–41 mutant alleles, nDf50 and gk262, exhibit 50% and 75% embryonic lethality respectively, whereas, wild type and hif-1(ia4) mutant embryos exhibit minimal lethality. At 0.5% O2, both mir-35–41 mutants approach 100% embryonic lethality (n = 176–242), whereas wild type and hif-1(ia4) mutant embryos exhibit 10% (n = 974) and 40% (n = 1132) lethality respectively. These data partially overlap with Table S1. (C) An increased percentage of nDf50 mutant embryos die younger in hypoxia than in normoxia. ‘Malformed’ refers to embryos with severe defects in their overall structure that do not permit stage identification. (D) Normoxic and hypoxic lethality of nDf50 mutant embryos is rescued by transgenic expression of mir-35. The sequence used to rescue mir-35 is shown as a green line in (A). n = 96–125. # refers to independent transgenic lines. Contingency table values are presented and Fischer exact test applied for statistical evaluation. ***≤0.001, ****≤0.0001. n.s. = not significant.
