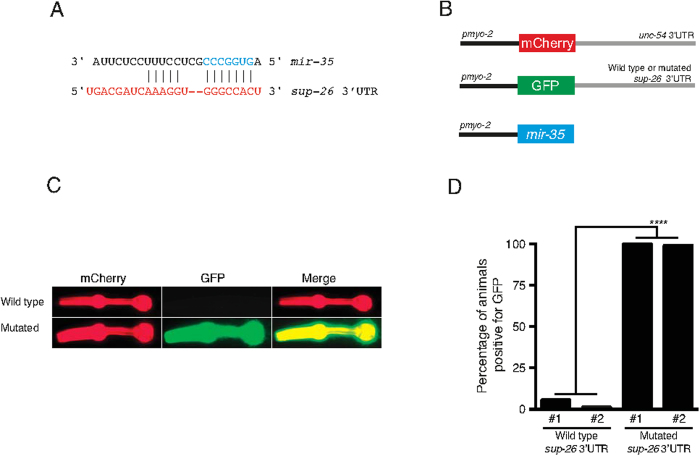
Figure 5. sup-26 is a mir-35–41 target and is required for hypoxic survival.
(A) The C. elegans sup-26 3’ UTR contains a single mir-35 family binding site (only mir-35 is shown). The mir-35 seed sequence is shown in blue and the predicted sup-26 3’UTR is shown in red. (B) Sensor experiment constructs. mir-35 was expressed in the pharynx together with a RFP reporter controlled by the unregulated unc-54 3’UTR and a GFP reporter controlled by the sup-26 3’UTR (wild type or mir-35 binding site mutated). The mir-35 binding site in the sup-26 3’UTR was mutated from CCCGGUG to CCatGgG to prevent binding of mir-35 family miRNAs. (C) Representative picture of the sensor experiment results. mir-35 downregulates GFP expression (sup-26 3’UTR) and not the RFP sensor (control 3’UTR). Regulation via the sup-26 3’UTR is dependent on the mir-35 binding site. (D) Quantification of the sensor experiment results. n > 50. Fischer exact test was used for statistical evaluation. # refers to independent transgenic lines. ****≤0.0001.