Abstract
Introduction
Accurate knowledge of O6-methylguanine methyltransferase (MGMT) gene promoter subtype in patients with glioblastoma (GBM) is important for treatment. However, this test is not always available. Pre-operative diffusion MRI (dMRI) can be used to probe tumour biology using the apparent diffusion coefficient (ADC); however, its ability to act as a surrogate to predict MGMT status has shown mixed results. We investigated whether this was due to variations in the method used to analyse ADC.
Methods
We undertook a retrospective study of 32 patients with GBM who had MGMT status measured. Matching pre-operative MRI data were used to calculate the ADC within contrast enhancing regions of tumour. The relationship between ADC and MGMT was examined using two published ADC methods.
Results
A strong trend between a measure of ‘minimum ADC’ and methylation status was seen. An elevated minimum ADC was more likely in the methylated compared to the unmethylated MGMT group (U = 56, P = 0.0561). In contrast, utilising a two-mixture model histogram approach, a significant reduction in mean measure of the ‘low ADC’ component within the histogram was associated with an MGMT promoter methylation subtype (P < 0.0246).
Conclusion
This study shows that within the same patient cohort, the method selected to analyse ADC measures has a significant bearing on the use of that metric as a surrogate marker of MGMT status. Thus for dMRI data to be clinically useful, consistent methods of data analysis need to be established prior to establishing any relationship with genetic or epigenetic profiling.
Keywords: ADC, diffusion MRI, glioblastoma, MGMT
Introduction
Currently, the median survival time for patients diagnosed with glioblastoma (GBM) is less than 15 months and patient treatment remains a significant clinical challenge.1 Recent literature has demonstrated the importance of the epigenetic status of the primary tumour, with methylation of the O6-methylguanine methyltransferase (MGMT) gene promoter improving survival.2–5 However, this MGMT status information is only available if patients have a resection, and, depending on the proximity and the technicality of the laboratory, is unavailable until after surgery and often only available well into treatment. There has been other work suggesting a detailed molecular profiling in GBM, in addition to MGMT status, codes for benefit from specific therapies.6,7 Thus earlier knowledge of molecular status could help more appropriate decisions to be made on treatment and management. Recently, it has been suggested that the pre-operative identification of magnetic resonance imaging (MRI) and imaging correlates of these specific molecular profiles is a plausible methodology to provide more timely, accurate and individualised treatment and management.8,9
As the most significant epigenetic profile on clinical outcomes in GBM to date, imaging surrogates of the MGMT promoter are of particular relevance. MGMT is an enzyme-like DNA protein which confers cellular resistance to alkylating agents effective in GBM such as temozolomide (TMZ)1 by removing the methyl adduct from the O6-position of guanine and subsequently preventing lethal DNA cross-link formation.10 Interestingly, it has also been suggested that the survival benefit of MGMT promoter methylation is not limited to patients treated with radiation therapy and TMZ; prolonged survival has also been reported in patients with methylated MGMT promoters irrespective of treatment modality.2 Consequently, MGMT promoter methylation status has been widely accepted as a powerful predictive and prognostic biomarker for patients with GBM.3–5
Due to the ability of diffusion MRI to probe tumour physiology on a macroscopic scale, a number of studies have reported imaging markers of MGMT promoter methylation status relating to quantitative indices such as the apparent diffusion coefficient (ADC). The rationale behind the use of this metric is based on the premise that tumour cellularity is inversely related to the ADC, that is, tumoural regions with low ADC correspond to areas of high cellularity.11 This assumption still requires further validation.7,12,13 Mean14 and minimum ADC values15,16 within the enhancing region on contrast-enhancing (CE) T1-weighted MRI have been reported to be elevated in GBM patients with methylated MGMT promoters whilst other studies employing histogram-based analyses have reported lower ADC associated with methylated MGMT gene promoter within the enhancing region.17 Given these previously reported opposing associations, we aimed to investigate the basis for the discrepancy in findings for the ADC and MGMT methylation association by analysing data using both minimum ADC and a two-mixture model histogram approach.18
Methods
The Institutional Ethics Review Board approved the study and written informed consent was obtained from each participant.
Patients
Data, collected during 2009–2012, from 32 patients (24 males, age range 38–69 years) with histopathologically confirmed high-grade brain tumour (WHO grade IV) were retrospectively analysed in this study. The patients were selected for inclusion on the basis of (1) known MGMT promoter methylation status, which was ascertained from tumour tissue obtained at resection, and (2) the availability of pre-operative CE, diffusion-weighted (DWI) and fluid-attenuated inversion recovery (FLAIR) MRI data. Brain extraction was employed before segmentation using the FSL (FreeSurfer Library) BET (Brain Extraction Tool).
Imaging protocols
MRI studies were acquired within 48 h before tumour resection. MRI scans were acquired using a 3T Siemens TimTrio (Siemens, Erlangen, Germany). Routine diagnostic scans were supplemented with a CE T1-weighted MRI acquired with a Magnetisation-prepared Rapid Acquisition Gradient-echo (MPRAGE) sequence with the following parameters (FOV 24 × 25.6 × 17.6 cm, TR/TE/TI 2300/2.26/900 msec, flip angle of 9°, 1 mm isotropic resolution). Images were acquired before and after administration of a gadolinium-based contrast agent (Gadovist®; Bayer HealthCare Pharmaceuticals, Sydney, Australia). DWI images were acquired in the axial plane using a spin-echo echo-planar sequence with diffusion gradient encoding in three orthogonal directions. The sequence parameters used (TR/TE 4500/91 msec), employed five averages and a maximum b value of 1000 sec/mm2. The image resolution was 1.1 × 1.1 ×5 mm. The diffusion scan was acquired before administration of the contrast agent.
Molecular analysis
Molecular analyses were performed on freshly frozen tissue obtained during tumour resection. MGMT promoter methylation status was assessed by methylation-specific PCR (Qiagen, Valencia, CA) as per the assay manufacturer's instructions.
Image processing
The quantitative image analysis was performed in a blinded manner using image-processing software tools available from the University of Oxford FMRIB Centre software library (version 4.0, http://www.fmrib.ox.ac.uk/). In this manner the ADC images were registered to each patients corresponding CE T1-weighted MRI image using a linear (affine) registration.19 The CE mask was extracted using the Automated Segmentation Tool within FMRIB employing a three-class segmentation model.20 This CE mask was then transformed to the corresponding, registered ADC map as shown in Figure1. For each patient, the accuracy of CE mask segmentation and registration to the ADC map was carefully visually assessed and manually corrected if required. For extracting the region of contrast enhancement, we compared the segmentation results using both three and four classes.
Figure 1.
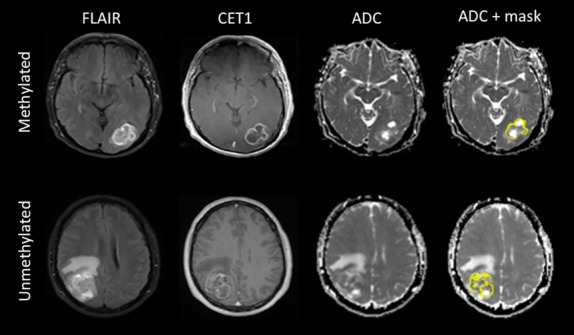
Defining the contrast enhancing mask for ADC analysis. Images include representative pre-operative FLAIR CET1 MRI, ADC and ADC and mask images from two patients with newly diagnosed primary glioblastoma with methylated O6-methylguanine-methyltransferase (MGMT) gene promoter (top) and unmethylated MGMT gene promoter (bottom). Associated masks highlighting the CE defined tumour region are given in yellow superimposed on the ADC maps. FLAIR, fluid-attenuated inversion recovery; CET1, contrast-enhancing T1-weighted; ADC, apparent diffusion coefficient.
ADC correlates
Two analysis approaches were employed to generate ADC values for correlation with MGMT promoter methylation status. First, the minimum ADC value within the CE portion of the tumour was determined using FMRIB statistical tools and expressed as a ratio to the minimum ADC value in a manually segmented region of interest within contra-lateral normal appearing brain parenchyma. A similar analysis strategy was employed by Moon et al.,15 who reported significantly higher ADC values were associated with MGMT methylation subtype. In contrast, a ‘two-mixture normal distribution’ histogram analysis and curve fitting of the ADC values within the mask were also performed, with mean values for the lower ADC distribution used for correlation with MGMT promoter methylation.18 This analysis strategy was employed by Pope et al.,17 who reported significantly lower ADC values associated with MGMT methylation subtype. It is proposed that the histogram approach is less biased towards ADC outlier values, which may occur due to possible registration error between the CE T1-weighted MRI and ADC maps.18 Representative CE masks delineated on registered CE T1-weighted MRI and ADC maps along with a two-mixture distribution histogram for the ADC indices are given in Figure2. ADC maps generated by the standard Siemens acquisition and reconstruction algorithms were used.
Figure 2.
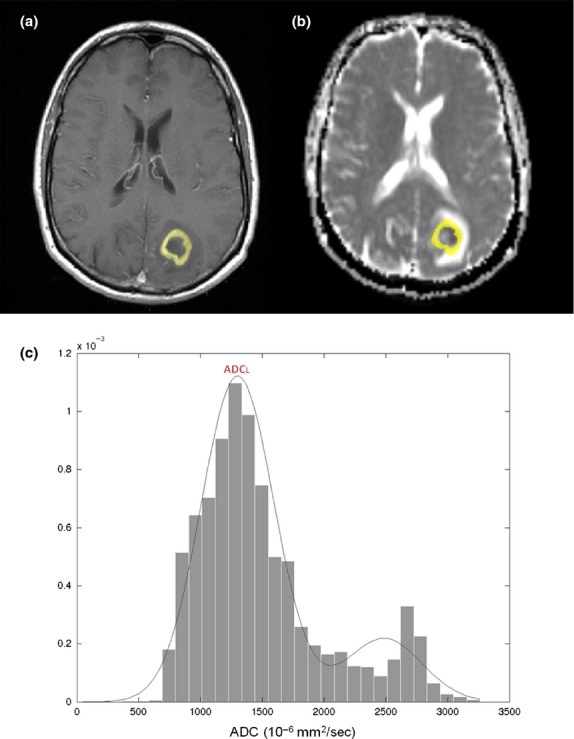
Generation of apparent diffusion coefficient (ADC) histograms and associated curves for data analysis. Representative images showing a contrast enhancement mask (yellow) overlaid on a contrast enhancing (CE) T1-weighted MRI (A) and registered ADC map (B). The corresponding histogram and curve fitting of ADC values within the CE mask is given in (C). The mean ADC values for the lower distribution (ADCL) were used to determine the association between ADC induces and MGMT promoter methylation status. O6-methylguanine-methyltransferase (MGMT).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA). Data were assessed for normality using the Shapiro–Wilk test and for homogeneity of variance prior to statistical analysis. Where data were normally distributed, parametric Student's t-tests were performed to determine whether there were any significant differences between imaging features and MGMT subtypes. Where the quantitative imaging data did not fit a normal distribution, non-parametric Mann–Whitney U-tests were used. Differences were considered statistically significant at P < 0.050.
Results
Patient demographics along with DW imaging metrics for the various MGMT methylation tissue signatures are given in Table1. In our cohort, 11 of the 32 patients had tumours with methylation of MGMT promoter (34%). There was no significant difference between the groups in age or gender, although there was a tendency for males to have unmethylated tumours. For extracting the region of contrast enhancement, we compared the segmentation results using both three and four classes. On the CE T1 images we found a three-class model yielded the best results for the entire cohort. With a three-class model (normal tissue, necrosis and contrast enhanced tissue), some cerebrospinal fluid was found to be classified as CE tissue.
Table 1.
Patient demographics and image features
| O6-methylguanine-methyltransferase promoter methylation status | P value | ||
|---|---|---|---|
| Methylated (n = 11) | Unmethylated (n = 21) | ||
| Age at diagnosis | |||
| mean (SD) | 57.54 (9.09) | 57.71 (9.49) | 0.961 |
| range | 38–68 | 37–69 | |
| Male:female | 6:5 | 18:3 | 0.087 |
| Image metrics mean (SD) | |||
| ADCmin | 1.236 (0.295) | 0.990 (0.289) | 0.0561 |
| ADCL (10−6 mm2/sec) | 1001 (95.10) | 1142 (221.0) | 0.0246 |
ADCmin, minimum apparent diffusion coefficient; ADCL, mean of the lowest distribution of apparent diffusion coefficient measure generated from the two-mixture model histogram approach.
With regard to ADC–MGMT promoter correlations, employing the use of the measure of minimum ADC, we found a strong trend towards the ADCmin being elevated within the methylated patient group compared to the unmethylated MGMT group (U = 56, P = 0.0561). Interestingly, utilising the two-mixture normal distribution histogram analysis, we found that the mean ADCL was significantly lower in the methylated MGMT patient group than in the unmethylated patient group, (t = 2.385, P < 0.0242). Box-whisker plots showing these distributions are given in Figure3. These contrasting results, obtained for the same patient cohort, highlight the varying effect different analysis strategies can have on the association between ADC indices and MGMT promoter methylation status.
Figure 3.

Box-whisker plots for imaging metrics relating to O6-methylguanine-methyltransferase (MGMT) methylation status. Plots showing the distribution (mean and standard deviation) for the diffusion-weighted imaging metrics based on MGMT promoter methylation status. The box-whisker plots represent the ADCmin (left) and ADCL (right). *Significant difference (P < 0.05). ADCmin, minimum apparent diffusion coefficient; ADCL, mean of the lowest distribution of the apparent diffusion coefficient measure generated from the two-mixture model histogram approach.
Discussion
In this study we have demonstrated that, within the same patient cohort, the association between ADC indices and MGMT promoter methylation status is dependent on the choice of analysis method. This finding helps to explain the contrasting reports in the literature regarding the use of minimum ADC values15,16 or histogram-based-derived measures17 to identify surrogate markers of MGMT subtype. As the histogram-based approach is less susceptible to outlier values caused by possible registration error, we believe that the histogram approach maybe a more robust method for determining MGMT promoter methylation status.
The aim of this study was to highlight that the relationship between diffusivity measures with MGMT status is dependent on the analysis method of choice. Currently, the widely accepted view is that low ADC values in newly diagnosed GBM are associated with a higher degree of tissue cellularity.15–17 However, from this study we cannot confirm whether reduced diffusivity is in fact due to an increase in cellular density11,13 and thus it is not clear whether increased cellularity is associated with methylated MGMT tumour subtypes. The relationship between such biological features needs to be explored in future studies.
There were a number of limitations of this study; firstly, it was a retrospective study with a relatively small sample size. Furthermore, we only assessed the degree of MGMT methylation from the resected tissue samples, making it difficult to speculate on possible interactions between other important factors such as tissue cellularity or other genetic and epigenetic parameters. For the ADC analysis, we analysed only the CE portion of the tumour as defined by semi-automated segmentation of the CE T1-weighted MRI image. This strategy was employed to match previously described analysis methods.18–23
It is also possible that non-linear registration of ADC to CE T1 images may have provided improved image fusion. However, after comparing both rigid and non-rigid registrations for the entire patient cohort, in terms of excessive warping of key anatomical features, we found that affine registration gave adequate results in all cases.
Conclusions
The findings of this study corroborate previous reports that elevated minimum ADC values and/or lower mean ADC values as determined by histogram analysis of DW images is associated with MGMT promoter methylation status. The current findings indicate that care should be taken when interpreting the link between diffusivity measures and genetic factors as the association is highly dependent of the method of ADC analysis. Overall, these findings provide a basis for future research into predictive and prognostic imaging biomarkers of genetic profiles in GBM. The development of sensitive and non-invasive imaging biomarkers could significantly help to further improve treatment planning and subsequently overall survival in patients with GBM.
Conflict of Interest
The authors declare no conflict of interest.
References
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Eng J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Brandes AA, Tosoni A, Franceschi E, et al. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J Clin Oncol. 2009;27:1275–9. doi: 10.1200/JCO.2008.19.4969. [DOI] [PubMed] [Google Scholar]
- Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–31. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- Chi AS, Batchelor TT, Dias-Santagata D, et al. Prospective, high-throughput molecular profiling of human gliomas. J Neurooncol. 2012;110:89–96. doi: 10.1007/s11060-012-0938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard D, Ibdaih A, Ducray F, et al. Primary brain tumors in adults. Lancet. 2012;379:1984–96. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. PNAS. 2008;105:5213–8. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas RF, Philips JJ, Parvataneni R, et al. Regional variation in histopathological features of tumor specimens from treatment-naïve glioblastoma correlates with anatomic and physiological MR imaging. Neuro-Oncol. 2012;14:942–54. doi: 10.1093/neuonc/nos128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Eng J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Cha S. Update on brain tumor imaging: from anatomy to physiology. AJNR Am J Neuroradiol. 2006;27:475–87. [PMC free article] [PubMed] [Google Scholar]
- Higano S, Yun X, Kumabe T, et al. Malignant astrocytic tumors: clinical importance of apparent diffusion coefficient in prediction of grade and prognosis. Radiology. 2006;241:839–46. doi: 10.1148/radiol.2413051276. [DOI] [PubMed] [Google Scholar]
- Rose S, Fay M, Thomas P, et al. Correlation of MRI-derived apparent diffusion coefficients in newly diagnosed gliomas with [18F]-fluoro-L-dopa PET: what are we really measuring with minimum ADC? AJNR Am J Neuroradiol. 2013;34:758–64. doi: 10.3174/ajnr.A3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo L, Choi SH, Park CK, et al. Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semi-quantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme. J Magn Reson Imaging. 2012;2:351–8. doi: 10.1002/jmri.23838. [DOI] [PubMed] [Google Scholar]
- Moon WJ, Choi JW, Roh HG, et al. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology. 2012;54:555–63. doi: 10.1007/s00234-011-0947-y. [DOI] [PubMed] [Google Scholar]
- Romano A, Calabria LF, Tavanti F, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol. 2012;23:513–20. doi: 10.1007/s00330-012-2601-4. [DOI] [PubMed] [Google Scholar]
- Pope WB, Lai A, Mehta R, et al. Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol. 2011;32:882–9. doi: 10.3174/ajnr.A2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WB, Kim HJ, Huo J, et al. Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology. 2009;252:182–9. doi: 10.1148/radiol.2521081534. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–56. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Al SA, Buckley R, McHenery C, et al. Distinguishing recurrent primary brain tumor from radiation injury: a preliminary study using a susceptibility-weighted MR imaging-guided apparent diffusion coefficient analysis strategy. AJNR Am J Neuroradiol. 2010;31:1049–54. doi: 10.3174/ajnr.A2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabycz S, Roldan G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage. 2010;49:1398–405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- Ellingson BM, Cloughesy TF, Pope WB, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. NeuroImage. 2012;59:908–16. doi: 10.1016/j.neuroimage.2011.09.076. [DOI] [PubMed] [Google Scholar]
- Ellingson BM, Lai A, Pope WB, et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol. 2013;34:533–40. doi: 10.3174/ajnr.A3253. [DOI] [PMC free article] [PubMed] [Google Scholar]


