Abstract
Excessive accumulation of nickel (Ni) can be toxic to plants. In Arabidopsis thaliana, the Fe2+ transporter, iron (Fe)-regulated transporter1 (IRT1), mediates Fe uptake and also implicates in Ni2+ uptake at roots; however, the underlying mechanism of Ni2+ uptake and accumulation remains unelucidated. In the present study, we found that zinc (Zn) deficient conditions resulted in increased accumulation of Ni in plants, particularly in roots, in A. thaliana. In order to elucidate the underlying mechanisms of Ni uptake correlating zinc condition, we traced 63Ni isotope in response to Zn and found that (i) Zn deficiency induces short-term Ni2+ absorption and (ii) Zn2+ inhibits Ni2+ uptake, suggesting competitive uptake between Ni and Zn. Furthermore, the Zrt/Irt-like protein 3 (ZIP3)-defective mutant with an elevated Zn-deficient response exhibited higher Ni accumulation than the wild type, further supporting that the response to Zn deficiency induces Ni accumulation. Previously, expression profile study demonstrated that IRT1 expression is not inducible by Zn deficiency. In the present study, we found increased Ni accumulation in IRT1-null mutant under Zn deficiency in agar culture. These suggest that Zn deficiency induces Ni accumulation in an IRT1-independen manner. The present study revealed that Ni accumulation is inducible in response to Zn deficiency, which may be attributable to a Zn uptake transporter induced by Zn deficiency.
Keywords: Arabidopsis thaliana, nickel, transporter, zinc
1. Introduction
Nickel (Ni) is known to be an essential nutrient for higher plants [1,2]; however, excessive amounts of Ni can be toxic [3]. In Ni-contaminated areas, agricultural crops exhibit Ni-induced impairments (e.g., chlorosis and leaf deformities), and the yields are reduced [4,5,6]. Thus, Ni phytotoxicity is one of the problems that limit agricultural production.
To elucidate the mechanism that underlies Ni phytotoxicity, we studied the molecular mechanism of Ni accumulation in plants using Arabidopsis thaliana as the model. We demonstrated that excess Ni is absorbed by the iron (Fe) uptake system in A. thaliana, which is associated with iron-regulated transporter 1 (IRT1), the primary Fe uptake transporter in roots [7]. Furthermore, we revealed that excess Ni accumulation induces IRT1 expression, which is associated with a disorder of Fe homeostasis, suggesting that Ni accumulation further accelerates Ni accumulation by IRT1 [8].
In our previous study, Ni accumulation was greatly decreased by defects in IRT1 under Fe-deficient conditions, whereas under Fe-sufficient conditions the reduction rate fell significantly, yet excess Ni still accumulated in the mutants, which exhibited Ni phytotoxicity [7]. This indicates that an additional pathway participates in the mediation of Ni accumulation in roots. Physiological evidence indicates that Ni2+ uptake competes with the magnesium (Mg) divalent ion in algae [9], barley [10], and spinach [11]. Further, members of the Arabidopsis MRS/MGT family of Mg2+ transporters have been shown to exhibit Ni2+ uptake activities in yeast [12,13]. These reports suggest that the Mg2+ uptake system is also a pathway for Ni2+ uptake. Furthermore, a previous study suggested that Ni2+ is absorbed by uptake systems for divalent heavy metal ions (e.g., Zn2+, Cu2+) in soybean [14]. Several studies have shown that some members of the Zrt/Irt-like protein (ZIP) family, which are the homologs of IRT1, are active in the transport of these heavy metal ions [15,16,17], suggesting that Ni2+ is competitively absorbed by the uptake systems of these divalent cations via ZIP transporters other than IRT1.
In the present study, we found that Ni accumulation increased in A. thaliana plants under Zn deficiency in hydroponics. A tracer assay revealed that the 63Ni absorption activity was higher in Zn-deficient plants than in the Zn-sufficient plants, showing that the Ni2+ uptake activity is upregulated by Zn deficiency. A zip3 mutant with an elevated Zn-deficient response exhibited increased Ni accumulation in roots compared with that in the wild type, possibly suggesting that the Zn-deficient response increases Ni accumulation. Furthermore, previous studies and our data suggest that the induction of Ni accumulation by Zn deficiency is attributable to an unknown molecular mechanism independent of IRT1.
2. Results
2.1. Ni Accumulation under Zn Deficiency in Hydroponics
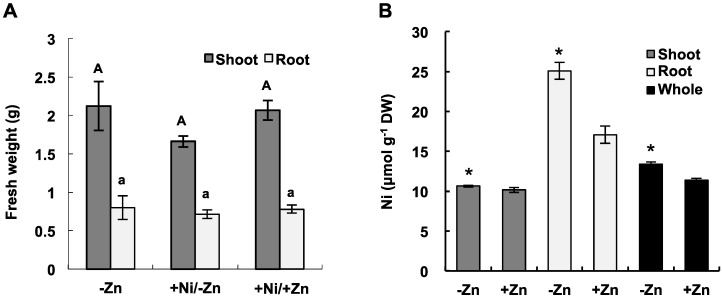
To investigate the effects of Zn deficiency on Ni accumulation in A. thaliana, growth rates and Ni accumulation were determined for 4-week old plants exposed to 25 μmol·L−1 NiCl2 under Zn-sufficient (5 μmol·L−1) or -deficient (0 μmol·L−1) conditions for 7 days in a hydroponic culture. As an additional control measure, plants were grown in the Zn-deficient conditions without Ni exposure. There was no statistically significant difference in growth between treatments (Figure 1A). Ni concentrations in the roots were 40% higher under the Zn-deficient conditions than those under the Zn-sufficient conditions (Figure 1B). A slight increase in Ni concentrations was observed in the shoots under the Zn-deficient conditions; however, Ni concentrations in whole plants were significantly higher under the Zn-deficient conditions than under the Zn-sufficient conditions.
Figure 1.
Ni accumulation in plants grown under Zn-sufficient or -deficient conditions. Four-week-old plants were exposed to 25 μmol·L−1 NiCl2 under Zn-sufficient (5 μmol·L−1) or -deficient (0 μmol·L−1) conditions for 7 days in a hydroponic culture. (A) Fresh weight. The same letters in each series (uppercase for shoots and lowercase for roots) indicate no significant difference (p ≥ 0.05, Steel-Dwass test) between treatments; (B) Ni concentrations in the shoots, roots, and whole plants. Asterisks denote significant differences (p < 0.05, Wilcoxon-Mann-Whitney test) between the Zn-sufficient and -deficient conditions for each tissue. The values represent means ± SD based on four independent experiments.
2.2. Short-Term 63Ni Absorption in Zn-Deficient/Sufficient Plants
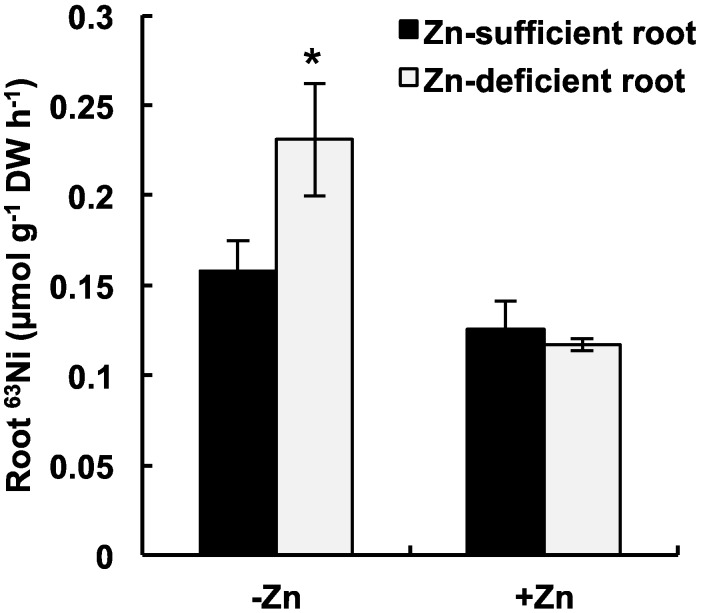
Ni concentration in roots represents the sum of the amount of Ni absorbed into the cell and the amount of Ni adsorbed onto the cell wall. On the cell wall, metal ions bind loosely to negatively charged sites, and the metals bound to the cell wall are readily removed by washing them with solutions containing competitive cations [11,18,19]. The proportion of Ni2+ among the total metal ions in the hydroponic solutions used in the above experiments was 0.0738% in both the Zn-absent and -present conditions. There was probably no difference in the amount of Ni fraction adsorbed in the different Zn conditions; therefore, we assumed that the increased Ni accumulation in roots under the Zn-deficient conditions represented the increased Ni absorption by the cells. Cataldo et al. [14] have suggested that Ni2+ and Zn2+ partially share the same uptake transporter in soybean. This could also be the case in A. thaliana, where Ni2+ was readily absorbed in the absence of competitive ions in the present study. Moreover, it is possible that the Ni2+ uptake activity of the roots was enhanced in response to Zn deficiency. To assess these possibilities, we performed a short-term Ni2+ uptake experiment using 63Ni-labeled NiCl2 (63NiCl2) and compared difference in Ni2+ uptake activities between Zn-deficient and -sufficient plants. One-month-old plants were grown under Zn-deficient (0 μmol·L−1) or -sufficient (5 μmol·L−1) conditions for 4 days in a hydroponic culture, and 63Ni uptake was determined with 5 μmol·L−1 63NiCl2 in the presence of Zn (5 μmol·L−1) or absence of Zn for 1 h. Our preliminary experiments revealed that 63Ni was almost undetectable in the shoots using this assay system; therefore, Ni uptake by plants was evaluated on the basis of 63Ni accumulation in the roots. To consider only the absorbed 63Ni, the apoplastic 63Ni was removed by incubating it in a wash solution containing 5 mmol·L−1 Ca(NO3)2 and 5 μmol·L−1 cold NiCl2, according to the established methods [11,18,19]. Statistical comparisons were made on 63Ni accumulation between the Zn-sufficient and -deficient roots in each Zn-present and -absent condition. Under the Zn-absent condition, 63Ni accumulation in the Zn-deficient roots was significantly higher (by 1.5-fold) than that in the Zn-sufficient roots (Figure 2), indicating that the 63Ni absorption activity was induced in the Zn-deficient roots, as we had expected. But, there was no significant difference in 63Ni accumulation between the Zn-sufficient and -deficient roots in the Zn-present condition. The addition of Zn to Zn-present and Zn-absent condition caused a significant decrease in Ni accumulation for both conditions (Wilcoxon-Mann-Whitney test, p < 0.05). This is suggesting that Ni2+ uptake is competitively inhibited by Zn2+, as observed in soybean.
Figure 2.
Short-term 63Ni uptake by plants. Plants grown under Zn-sufficient (5 μmol·L−1) or -deficient (0 μmol·L−1) conditions for 4 days were incubated in hydroponic solutions containing 5 μmol·L−1 63NiCl2 in the presence of Zn (5 μmol·L−1 ZnCl2) or absence of Zn (0 μmol·L−1) for 1 h. Asterisks denote significant differences (p < 0.05, Wilcoxon-Mann-Whitney test) between the Zn-sufficient and -deficient roots, and the values represent means ± SD based on four independent experiments.
2.3. Ni Accumulation in a ZIP3 Mutant
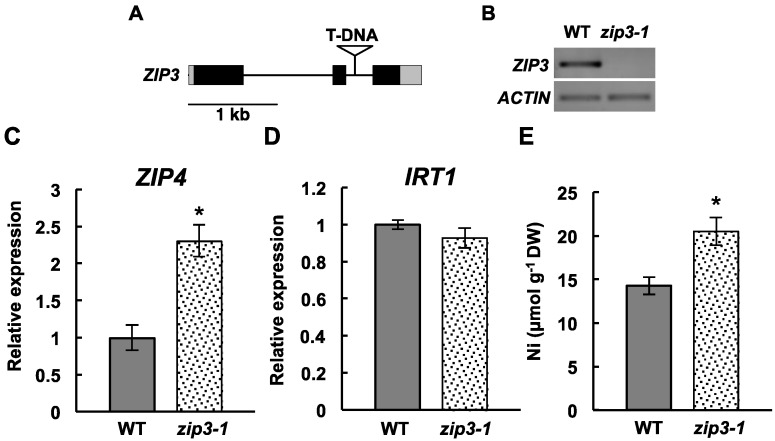
ZIP3 is a member of the ZIP family, and it has been shown to be highly induced by Zn deficiency in the roots of A. thaliana [16,20]. It was also found that ZIP3 complemented the Zn2+-uptake transporter of yeasts [15]. Therefore, it has been suggested that ZIP3 is involved in Zn nutrition of the roots. We obtained a T-DNA mutant zip3-1, which carries a T-DNA insertion in the second intron of ZIP3 (Figure 3A). The expression of the full length of ZIP3 was undetectable in zip3-1 (Figure 3B), confirming that this strain is defective in ZIP3 expression. Four-week-old plants of the zip3-1 mutant and wild type were grown under Zn-sufficient conditions (5 µM ZnCl2) for 1 week in hydroponics, and we examined ZIP4 expression, which is a well-known indicator of Zn deficiency [21]. ZIP4 expression increased more than twofold in zip3-1 compared with that in Col-0 (Figure 3C), indicating that the Zn-deficient response was elevated in zip3-1, even under Zn-sufficient conditions. There was no significant difference in IRT1 expression between the zip3-1 mutant and the wild type (Figure 3D). Next, we exposed 4-week-old zip3-1 mutant plants and wild type plants to 25 µM NiCl2 under Zn-sufficient conditions (5 µM ZnCl2) for 1 week and determined Ni accumulation in the roots. Ni accumulation increased by 1.5 fold in zip3-1 compared with that in the wild type (Figure 3E). This result is suggesting that Ni accumulation can be increased in a mutant that exhibits an endogenously elevated Zn-deficient response, which is supporting the hypothesis that Ni accumulation increases in plants in response to Zn deficiency.
Figure 3.
Ni accumulation in the Zrt/Irt-like protein 3 (ZIP3) mutant. (A) Scheme showing the T-DNA insertion site in zip3-1; (B) Semi-quantitative amplification of full-length CDS of ZIP3 in the roots of the wild type and zip3-1. The results shown were obtained after 30 cycles of amplification. Actin was amplified as an endogenous control; (C,D) Quantification of the expression levels of ZIP4, a well-known Zn-deficient response indicator and IRT1 in roots grown in Zn-sufficient condition (5 µM ZnCl2) for 1 week. The expression levels relative to EF1α are shown; (E) Ni accumulation in roots. Four-week-old plants were exposed to 25 μmol·L−1 NiCl2 under Zn-sufficient conditions (5 μmol·L−1 ZnCl2) for 1 week in hydroponic culture. The data represent means ± SD based on three (C,D) and four (E) independent experiments. Asterisks denote significant differences (p < 0.05, Wilcoxon-Mann-Whitney test) between zip3-1 and the wild type.
2.4. Ni Accumulation in the IRT1 Mutant
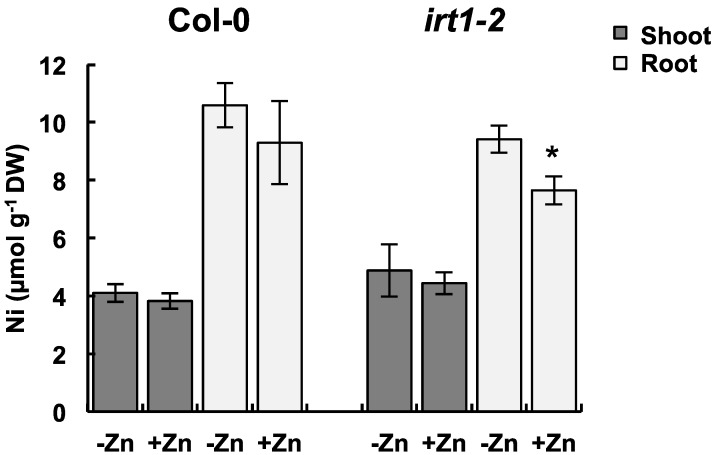
We also examined Ni accumulation in Zn-deficient and -sufficient conditions in an IRT1-defective mutant. We reported two independent IRT1-defective mutants in the Col-0 background [7]. But, here we used irt1-2, which has a null mutation in IRT1, to exclude the effect of IRT1 as much as possible. irt1-2 carries a T-DNA insertion in the open-reading frame of IRT1 and expresses a mutated IRT1 coding for a non-functional form [7]. Because the irt1-2 plants cannot grow in the normal hydroponic culture, we needed to use an agar plate containing sucrose to maintain irt1-2 growth as described in our previous report. As a result of preliminary test with Col-0, we confirmed that Zn accumulations in the shoots and roots of plants grown on the Zn-deficient plate were significantly decreased compared with those of plants grown on the Zn-sufficient plate (Figure S1). In Col-0 plants, the mean Ni concentration in roots was increased by 15% in the Zn-deficient conditions compared with that in the Zn-sufficient conditions, although not statistically significant (p = 0.3, Wilcoxon-Mann-Whitney test) (Figure 4), possibly suggesting that Ni accumulation is increased by Zn deficiency in agar culture. In irt1-2 plants, Ni accumulation in roots was significantly increased under the Zn-deficient conditions. These results indicate that the increased Ni accumulation by Zn deficiency is also observed in agar culture at least in irt1-2.
Figure 4.
Ni accumulation in iron-regulated transporter1-2 (irt1-2) under Zn-sufficient and -deficient conditions. One-week-old plants were grown on MGRL plates containing 25 μmol·L−1 NiCl2 and 0 or 5 μmol·L−1 ZnCl2 for 1 week. Asterisks denote significant differences (p < 0.05, Wilcoxon-Mann-Whitney test) between the Zn-sufficient and -deficient conditions. The values represent means ± SD based on four independent experiments.
3. Discussion
In this study, we showed that Ni accumulation was increased by Zn deficiency in A. thaliana. An increased Ni accumulation was clearly observed in the roots, whereas the increase was quite small in the shoots. In a previous study, no marked difference was noted in Ni accumulation in the shoots of the irt1 mutant and the wild type, although Ni accumulation in roots was much lower in irt1 [7]. Thus, it appears that the effect of increased or decreased Ni absorption in roots is not reflected by Ni accumulation in shoots. Therefore, we mainly focused on Ni accumulation in roots in the present study.
In a radioisotope tracer experiment, we assessed Ni2+ absorption in roots treated under Zn-deficient and -sufficient conditions and found that 63Ni absorption was higher in Zn-deficient roots, indicating that the Ni2+ absorption activity is induced in response to Zn deficiency in wild type plants. We also found that the zip3 mutant, which exhibits a constitutively elevated Zn-deficient response, accumulated higher levels of Ni than the wild type. This observation further supports that Ni2+ absorption is inducible in response to Zn deficiency. In the tracer assay, we also observed that the increased 63Ni absorption in Zn-deficient roots was considerably inhibited by adding Zn to the uptake solution, suggesting that Ni2+ can be competitively absorbed via a Zn2+ uptake pathway, which is similar to that found in soybean [14]. These findings may indicate that Ni2+ is absorbed via a Zn2+ uptake transporter induced by Zn deficiency in roots.
It is known that IRT1 is active in Zn2+ uptake [22,23]; however, its expression is not induced by Zn deficiency [16,20]. Further, the irt1 mutant exhibited an increased Ni accumulation under Zn-deficient conditions in agar culture in the present study. These are suggesting that the increased Ni accumulation caused by Zn deficiency occurs independently of IRT1. But, we note that further studies are necessary in order to conclude that Zn-deficient induced Ni accumulation observed in hydroponic culture is also independent of IRT1. IRT1 expression did not increase in the zip3 mutant, and thus IRT1 was probably not the cause of the increased Ni accumulation in the zip3 mutant.
The findings of the present study provide considerable insights to elucidate the molecular mechanisms of Ni accumulation in plants. In A. thaliana, it has been shown that two closely related members of the basic-region leucine-zipper family of transcription factors, bZIP19 and bZIP23, are responsible for the primary responses to Zn deficiency and that the bzip19 bzip23 double mutant is unable to induce some genes responsive to Zn deficiency [21]. In the present study, Ni accumulation caused by Zn deficiency could have been controlled by these transcription factors. Our data also suggest that Ni2+ is competitively taken up by a Zn2+ transporter. The transporter genes that play a role in Zn2+ uptake by A. thaliana roots are uncertain; however, as previously mentioned, several studies have shown that some ZIP family genes encode Zn2+ uptake transporters, which are induced by Zn deficiency [17,20,21]. Therefore, it is expected that other ZIP members will also have Ni2+ transport activities. Currently, we are investigating the possible involvement of other Arabidopsis ZIP genes in Ni2+ uptake.
4. Experimental Section
4.1. Analysis of Ni Accumulation in Plants Grown in Hydroponic Culture and Agar Plate Culture
The A. thaliana ecotype Col-0 was cultured in a hydroponic system, as described in our previous study [7]. Four-week-old plants were irrigated with a hydroponic solution containing 25 μmol·L−1 NiCl2 and 0 or 5 μmol·L−1 ZnCl2 and were cultured for 1 week. After exposure, the plants were rinsed with deionized water, and the fresh weights were determined. The plant parts were then dried at 70 °C for 3 days to determine the dry weight. We confirmed that the abundance of free Ni2+ in the hydroponic solution was >99% of the total Ni in both the Zn-sufficient and -deficient conditions using GEOCHEM-EZ [24].
Plants with irt1-2 (SALK_054554), which is a previously described null mutant of IRT1 in the Col-0 background [7], and the wild type were grown on MGRL-base agar plates, which comprised half-strength MGRL supplemented with 1.5% sucrose, 1.2% ultra-pure agar (Sigma-Aldrich Co., Tokyo, Japan), 5 mmol·L−1 MES (pH 5.5), and 50 μmol·L−1 FeNa-EDTA. One-week-old seedlings were transferred to agar plates, which were supplemented with 25 μmol·L−1 NiCl2 and 0 or 5 μmol·L−1 ZnCl2 and grown for 1 week. After exposure, the plants were sampled as described above.
The plant samples were digested with HNO3 and H2O2 in a heat block. The elemental analysis was performed using an inductively coupled plasma (ICP) atomic emission spectrometer (ICPS-7500; Shimadzu Co., Kyoto, Japan) or ICP mass spectrometer (SPQ9700; Hitachi High-Teck Science Co., Tokyo, Japan).
4.2. Short-Term Ni2+ Uptake Assay Using 63Ni Isotope
One-month-old plants cultured under hydroponic conditions were grown in hydroponic solutions containing 0 or 5 μmol·L−1 ZnCl2 for 4 days. Next, the roots were rinsed with deionized water and gently blotted, and the plants were irrigated with a hydroponic solution containing 5 μmol·L−1 63NiCl2 (2.16 µCi·mL−1, Nuclitec GmbH, Braunschweig, Germany) with or without 5 μmol·L−1 ZnCl2, and incubated in the growth chamber under the conditions described above. After 1 h, the plants were placed into wash solutions containing 5 mmol·L−1 Ca(NO3)2 and 5 μmol·L−1 NiCl2 for 30 min at room temperature. This step facilitated the desorption of 63Ni ions, which were adsorbed onto the root cell walls via the equilibrium effect, as previously described [11,18,19]. The 63Ni contents of the roots were determined using previously described procedures [7] with a liquid scintillation counter (LSC-5100; Hitachi Aloka Medical Ltd., Tokyo, Japan). The hydroponic culture conditions were the same as those described above.
4.3. Identification of the ZIP3-Defective Mutant and Gene Expression Analysis
The T-DNA mutant lines zip3-1 (CS870394) were acquired from the Syngenta Arabidopsis Insertion Library (SAIL) collection via the Arabidopsis Biological Resource Center (ABRC). Homozygous insertion mutants were identified using genomic polymerase chain reaction (PCR) with the following primers: 5'-TGA CTA ACA GAA CTA TAG AAG ACG CAT G-3' and 5'-CAT GCC ATT TAT TGT CGA TGA TGA CGA-3'.
The Col-0 and zip3-1 plants were grown in hydroponics for 4 weeks, as described above, and the plants were then transferred to Zn-sufficient conditions (5 μmol·L−1 ZnCl2) and cultured for 1 week. Total RNA was extracted from the roots, according to the method reported by Suzuki et al. [25]. Genomic DNA in the extracts was digested with DNase I (Takara Bio Inc., Shiga, Japan), and the first-strand cDNA was synthesized from the total RNA using the PrimeScript™ RT reagent kit (Takara Bio Inc.). The cDNA was used in the following semi-quantitative and quantitative RT-PCR assays.
Amplification of the full-length coding region of ZIP3 (AT2G32270) was conducted using the above mentioned gene-specific primers and cDNA corresponding to 5 ng of RNA. PrimerSTAR® HS DNA Polymerase (Takara Bio Inc.) was used for PCR. ACTIN (AT2G37620) was also amplified as an internal control with the following primers: 5'-AAT TGG GAT GAC ATG GAG AAG ATT TGG-3' and 5'-TGG AGT TAT AGG TGG TTT CAT GGA TAC-3'. The PCR products on the agarose gels were detected by staining with ethidium bromide.
cDNA corresponding to 5 ng of RNA was used for quantitative RT-PCR with SYBR Premix Ex Taq II (Takara Bio Inc.). The PCR reaction was performed with a Thermal Cycler Dice Real Time System II® (Takara Bio Inc.). For the thermal profile, instructions included in the manual provided with the PCR kit were followed. The specific primers used for ZIP4 (AT1G10970) were as follows: 5'-GGT TGT ATC CTC CAG GCT GAG T-3' and 5'-TGG TGT TGT TAC CGC GAA AA-3'. The specificities of the primers were confirmed by analyzing the dissociation curves and agarose gel electrophoresis of the PCR products. The specific primers for EF1α designed by Takano et al. [26] were used.
5. Conclusions
In the present study, we demonstrated that Zn deficiency induces Ni absorption activity in roots and Zn2+ inhibits Ni2+ uptake. We also found Ni accumulation can be increased in IRT1-null mutant. Taken together, our data suggest that Ni is absorbed by an unknown Zn transporter induced by Zn deficiency.
Acknowledgments
We thank Toru Fujiwara (The University of Tokyo) for technical support in an elemental analysis with ICP-MS. We also thank Myles M. Butler and Ai Kitazumi (The University of Maine) for critically reading and Enago (www.enago.jp) for the English language review. This work was supported, in part, by a Grant-in-Aid for JSPS Fellows 09J05716 (Sho Nishida), a Grant-in-Aid for Young Research (B) 18780045 and for Scientific Research (B) 24380038 (Takafumi Mizuno) from the Japan Society for the Promotion of Science.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/05/9420/s1.
Author Contributions
Sho Nishida designed the experiments. Sho Nishida, Chisato Tsuzuki, Aki Kato, and Junko Yoshida performed the experiments and analyzed data. Sho Nishida and Takafumi Mizuno have contributed in writing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Brown P.H., Welch R.M., Cary E.E. Nickel: A micronutrient essential for higher plants. Plant Physiol. 1987;85:801–803. doi: 10.1104/pp.85.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown P.H., Welch R.M., Madison J.T. Effect of nickel deficiency on soluble amino acid, and nitrogen levels in barley. Plant Soil. 1990;125:19–27. doi: 10.1007/BF00010740. [DOI] [Google Scholar]
- 3.Seregin I.V., Kozhevnikova A.D. Physiological role of nickel and its toxic effects on higher plants. Russ. J. Plant Physiol. 2006;53:257–277. doi: 10.1134/S1021443706020178. [DOI] [Google Scholar]
- 4.Temple P.J., Bisessar S. Uptake and toxicity of nickel and other metals in crops grown on soil contaminated by a nickel refinery. J. Plant Nutr. 1981;3:473–482. doi: 10.1080/01904168109362852. [DOI] [Google Scholar]
- 5.Frank R., Stonefield K.I., Suda P. Impact of nickel contamination on the production of vegetables on an organic soil, Ontario, Canada, 1980–1981. Sci. Total Environ. 1982;26:41–65. doi: 10.1016/0048-9697(82)90095-X. [DOI] [Google Scholar]
- 6.Bisessar S. Effects of lime on nickel uptake and toxicity in celery grown on muck soil contaminated by a nickel refinery. Sci. Total Environ. 1989;84:83–90. doi: 10.1016/0048-9697(89)90372-0. [DOI] [PubMed] [Google Scholar]
- 7.Nishida S., Tsuzuki C., Kato A., Aisu A., Yoshida J., Mizuno T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:1433–1442. doi: 10.1093/pcp/pcr089. [DOI] [PubMed] [Google Scholar]
- 8.Nishida S., Aisu A., Mizuno T. Induction of IRT1 by the nickel-induced iron-deficient response in Arabidopsis. Plant Signal. Behav. 2012;7:329–331. doi: 10.4161/psb.19263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worms I.A., Parthasarathy N., Wilkinson K.J. Ni uptake by a green alga. 1. Validation of equilibrium models for complexation effects. Environ. Sci. Technol. 2007;41:4258–4263. doi: 10.1021/es0630339. [DOI] [PubMed] [Google Scholar]
- 10.Lock K., van Eeckhout H., de Schamphelaere K.A., Criel P., Janssen C.R. Development of a biotic ligand model (BLM) predicting nickel toxicity to barley (Hordeum vulgare) Chemosphere. 2007;66:1346–1352. doi: 10.1016/j.chemosphere.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Degryse F., Shahbazi A., Verheyen L., Smolders E. Diffusion limitations in root uptake of cadmium and zinc, but not nickel, and resulting bias in the Michaelis constant. Plant Physiol. 2012;160:1097–1109. doi: 10.1104/pp.112.202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Tutone A.F., Drummond R.S., Gardner R.C., Luan S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell. 2001;13:2761–2775. doi: 10.1105/tpc.13.12.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L.G., Sokolov L.N., Yang Y.H., Li D.P., Ting J., Pandy G.K., Luan S. A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol. Plant. 2008;1:675–685. doi: 10.1093/mp/ssn031. [DOI] [PubMed] [Google Scholar]
- 14.Cataldo D.A., Garland T.R., Wildung R.E. Nickel in plants: I. Uptake kinetics using intact soybean seedlings. Plant Physiol. 1978;62:563–565. doi: 10.1104/pp.62.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grotz N., Fox T., Connolly E., Park W., Guerinot M.L., Eide D. Identification of a family of Zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wintz H., Fox T., Wu Y.Y., Feng V., Chen W., Chang H.S., Zhu T., Vulpe C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- 17.Milner M.J., Seamon J., Craft E., Kochian L.V. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 2013;64:369–381. doi: 10.1093/jxb/ers315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasat M.M., Baker A.J., Kochian L.V. Physiological characterization of root Zn2+ absorption and translocation to shoots in Zn hyperaccumulator and nonaccumulator species of Thlaspi. Plant Physiol. 1996;112:1715–1722. doi: 10.1104/pp.112.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer U., Smith R.D., Wenzel W.W., Raskin I., Salt D.E. The role of metal transport and tolerance in nickel hyperaccumulation by Thlaspi goesingense Hálácsy. Plant Physiol. 1997;115:1641–1650. doi: 10.1104/pp.115.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van de Mortel J.E., Almar Villanueva L., Schat H., Kwekkeboom J., Coughlan S., Moerland P.D., Ver Loren van Themaat E., Koornneef M., Aarts M.G. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006;142:1127–1147. doi: 10.1104/pp.106.082073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assunção A.G., Herrero E., Lin Y.F., Huettel B., Talukdar S., Smaczniak C., Immink R.G., van Eldik M., Fiers M., Schat H., et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA. 2010;107:10296–10301. doi: 10.1073/pnas.1004788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korshunova Y.O., Eide D., Clark W.G., Guerinot M.L., Pakrasi H.B. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999;40:37–44. doi: 10.1023/A:1026438615520. [DOI] [PubMed] [Google Scholar]
- 23.Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaff J.E., Schultz B.A., Craft E.J., Clark R.T., Kochian L.V. GEOCHEM-EZ: A chemical speciation program with greater power and flexibility. Plant Soil. 2010;330:207–214. doi: 10.1007/s11104-009-0193-9. [DOI] [Google Scholar]
- 25.Suzuki Y., Kawazu T., Koyama H. Arabidopsis RNA isolation. Biotechniques. 2004;37:542–544. doi: 10.2144/04374BM03. [DOI] [PubMed] [Google Scholar]
- 26.Takano J., Wada M., Ludewig U., Schaaf G., von Wirén N., Fujiwara T. The Arabidopsis major intrinsic protein NIP5; 1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell. 2006;18:1498–1509. doi: 10.1105/tpc.106.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.