Figure 6.
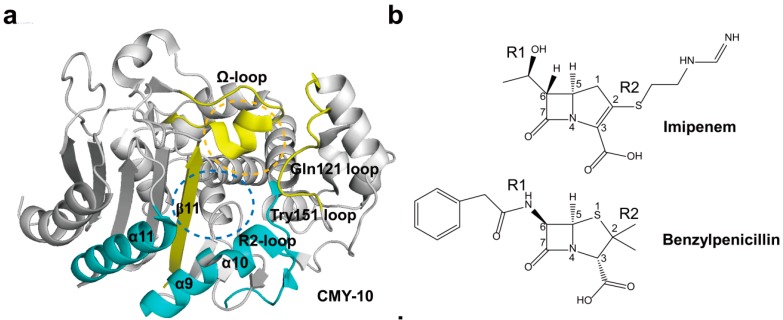
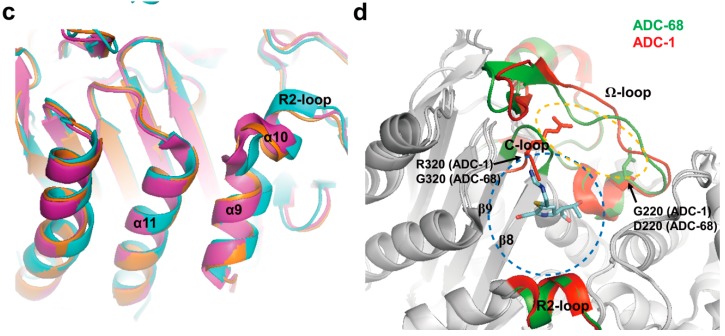
(a) Overall structure of CMY-10 (PDB entry 1ZKJ) is shown. The R1 subsite is surrounded by the Ω-loop, Gln121 loop, and β11 (in yellow). The R2 subsite is enclosed by the Tyr151 loop, α10 in the R2-loop, and α11 (in cyan). The R1 and R2 subsites are indicated as orange and blue dotted circles, respectively; (b) Schematic drawing of imipenem and benzylpenicillin is shown. The β-lactam nucleus is numbered. The R1 and R2 side chains located at the C6 and C2 positions of the β-lactam nucleus are labeled, respectively; (c) The displacement of α9 and α10 in CMY-10 is shown. CMY-10 (PDB entry 1ZKJ, cyan) was superposed with P99 β-lactamase (PDB entry 2BLT, orange) and GC1 β-lactamase (PDB entry 1GCE, magenta [89]). The R2-loop displays noticeable structural alterations: the R2-loop becomes flexible, and the shortened path of the connection loop between α10 and β11 induces the ~2.5 Å shift of α9 and α10 relative to the adjacent helix α11 in CMY-10 compared with both P99 and GC1 β-lactamases; (d) Superimposed complex of imipenem with ADC-68 and ADC-1. An AmpC complex with imipenem (PDB entry 1LL5) was superposed with ADC-1 (PDB entry 4NET) and ADC-68 (PDB entry 4QD4). ADC-68 and ADC-1 are represented as green and red ribbon diagrams, respectively. Imipenem is represented as cyan stick. C-loop (T318–F321) is positioned between β8 and β9. Ω-loop (G185–T229) is positioned between α6 and α8. R2-loop (E291–V309) is positioned between α9 and α10b. The R1 and R2 subsites are indicated as orange and blue dotted circles, respectively. R320 (ADC-1) and G320 (ADC-68) residues are located in C-loop and G220 (ADC-1) and D220 (ADC-68) residues are found in Ω-loop. Superpositions were performed using SSM Superpose [53] to align the complete chains. The structures of CMY-10 and ADC-68 were prepared using PyMOL [54] and data adapted from Kim et al. [80] and Jeon et al. [12], respectively.