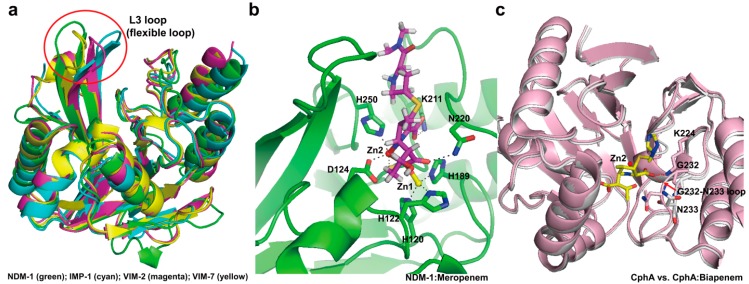
Figure 13.
(a) Comparison of the structural features of NDM-1 (PDB entry 3SPU, green) with IMP-1 (PDB entry 1DD6, cyan), VIM-2 (PDB entry 1KO2, magenta), and VIM-7 (PDB entry 2Y87, yellow), showing the significant differences in orientation of the L3 loop. L3 loops are indicated by the red circle; (b) Active site of the NDM-1 in complex with meropenem (PDB entry 4EYL, green) is shown. The residues (H120, H122, D124, H189, K211, N220, and H250; NDM-1 numbering) in the active-site cleft are shown as green sticks. Zinc coordination and hydrogen bonding in active site in NDM-1 are shown as dashed black lines. The hydrolyzed meropenem is shown in magenta. Zn1 and Zn2 are represented as yellow and red spheres, respectively; (c) The conformational change upon substrate binding is represented by superimposition of the wild-type CphA (PDB entry 1X8G, white) and CphA (N220G mutant):biapenem complex (PDB entry 1X8I, light pink). The residues (K224, G232, and N233; class B numbering) in the active-site cleft are shown as sticks. The movement (from open to closed position) of G232-N233 loop appears as a red arrow. The closed position is shown in CphA (N220G mutant):biapenem complex structure. The hydrogen bond interactions between the C3 carboxyl group of biapenem and two residues (K224 and N233) in the active site in CphA are shown as dashed red lines. The hydrolyzed biapenem and Zn2 are represented as yellow stick mode and red sphere, respectively. Superpositions were performed using SSM Superpose [53] to align the complete chains. The structures of NDM-1 and CphA were prepared using PyMOL [54] and data adapted from King et al. [149] and Garau et al. [154], respectively.