Figure 4.
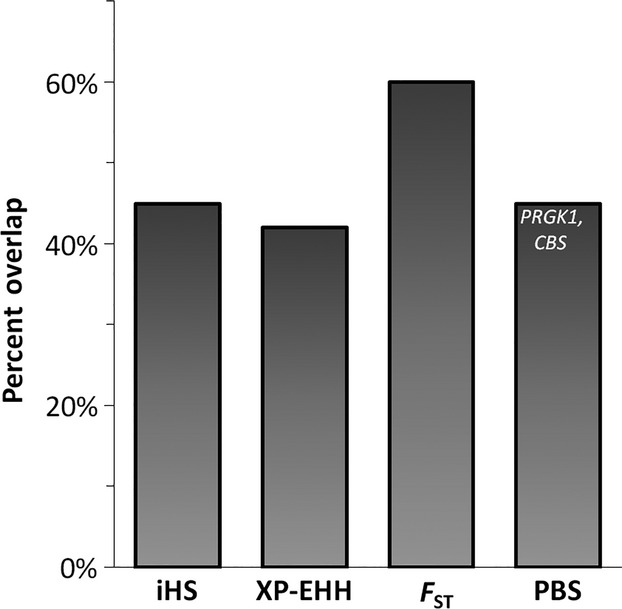
Overlap of the top 1% selection test results in Calchaquíes and with the top 5% in Collas. The overlap of windows highlighted by selection tests was the highest for FST with 60%. However, only two out of 213 candidate genes (PRKG1 and CBS) overlapped, both discovered using PBS. PRKG1 ranked higher in Calchaquíes with a PBS score of 0.836 (rank 5) than in Collas with a score of 0.693 (rank 52). Thus, allele frequencies were more diverged in Calchaquíes than in Collas. The PBS score is based on the same SNP (rs10740406) for both populations. CBS ranked lower in Calchaquíes (rank 48, PBS score 0.632) than in Collas (rank 2, PBS score 1.005), indicating a less pronounced allele differentiation in Calchaquíes in comparison to Collas for this gene. For CBS different SNPs within the same 100 kb window were highlighted in the two populations. CBS, cystathionine-beta-synthase; FST, Fixation Index; iHS, integrated Haplotype Score; PBS, Population Branch Score; PRKG1, protein kinase, cGMP-dependent, type I; XP-EHH, Cross Population Extended Haplotype Homozygosity test.
