Figure 4.
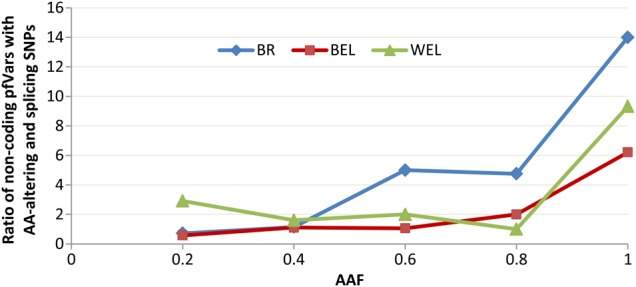
Ratio of non-coding putatively functional variants (pfVars) with AA altering and splicing types at different allele frequency bins within selective sweep regions. The non-coding pfVars included intergenic, intronic, UTR and down/upstream variants from most conserved elements, variants located within known ncRNAs and variants that were predicted to alter RNA secondary structure. The graph indicates that high-frequency variants are enriched with non-coding types. BR, BEL and WEL refer to broiler, brown egg layer and white egg layer groups, respectively.
