Abstract
Hepatitis C virus (HCV) often causes persistent infection, and is an important factor in the etiology of fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). There are no preventive or therapeutic vaccines available against HCV. Treatment strategies of HCV infection are likely to improve with recently discovered direct antiviral agents (DAAs). However, a proportion of patients still progress to liver failure and/or HCC despite having been cured of the infection. Thus, there is a need for early diagnosis and therapeutic modalities for HCV related end stage liver disease prevention. HCV genome does not integrate into its host genome, and has a predominantly cytoplasmic life cycle. Therefore, HCV mediated liver disease progression appears to involve indirect mechanisms from persistent infection of hepatocytes. Studying the underlying mechanisms of HCV mediated evasion of immune responses and liver disease progression is challenging due to the lack of a naturally susceptible small animal model. We and other investigators have used a number of experimental systems to investigate the mechanisms for establishment of chronic HCV infection and liver disease progression. HCV infection modulates immune systems. Further, HCV infection of primary human hepatocytes promotes growth, induces phenotypic changes, modulates epithelial mesenchymal transition (EMT) related genes, and generates tumor initiating stem-like cells (TISCs). HCV infection also modulates microRNAs (miRNAs), and influences growth by overriding normal death progression of primary human hepatocytes for disease pathogenesis. Understanding these ob-servations at the molecular level should aid in developing strategies for additional effective therapies against HCV mediated liver disease progression.
Keywords: HCV, DAAs, miRNAs, EMT, fibrosis, TISCs
Introduction
Hepatitis C virus (HCV) is an enveloped virus with a ~9.6 kb single-stranded RNA genome (Choo et al., 1989[33]), a member of the Flaviviridae family and genus Hepacivirus. HCV genome encodes a single polyprotein which is processed co-translationally into three structural and seven nonstructural (NS) polypeptides (Grakoui et al., 1993[64]; Tanji et al., 1994[182]; Ali et al., 2011[5]). HCV core protein forms the capsid, which is surrounded by a lipid bilayer containing the envelope glycoproteins, E1 and E2 on the external surface. These envelope glycoproteins are responsible for initiation of infection in a host cell. The nonstructural (NS) proteins coordinate the intracellular processes of the virus life cycle.
HCV is a major cause of chronic liver disease, with an estimated 180 million people infected worldwide. An important therapeutic advancement was achieved with the recent discovery of potent direct acting antiviral agents (DAAs) against HCV (Casey and Lee, 2013[30]; Au and Pockros, 2014[9]). Several clinical trials have shown various combinations of agents, including interferon-free regimens, to be highly effective in the clearance or sustained viral response (SVR) of chronic hepatitis C infection. However, significant challenges remain in deploying modern antivirals for patients with asymptomatic HCV infection and must be sought through screening programs. HCV infection particularly affects persons of low socio-economic status who have less access to health care. The very high cost of HCV treatment may also contribute to delays in patients being treated.
Majority of the infected patients (approximately 80 %) develop chronic infection and are at high risk for end stage liver disease progression to cirrhosis and hepatocellular carcinoma (HCC). HCC is a common cancer worldwide and accounts for ~5.6 % of all cancers. It is the fifth common cancer in the world and the third common cause of cancer death (Bosch et al., 2004[22]; Sherman, 2010[170]). The incidence of HCC is rising precipitously, primarily as a result of the increasing prevalence of chronic HCV infection (Kanwal et al., 2011[89]) and fatty liver disease in the United States (Nordenstedt et al., 2010[139]; Zhang and Friedman, 2012[202]). Liver fibrosis is strongly associated with HCC, since approximately 80-90 % of HCC cases are arising in cirrhotic livers (Seitz and Stickel, 2006[168]; Lok et al., 2009[110]). HCC development is also linked to alcoholic cirrhosis (Fattovich et al., 2004[50]), nonalcoholic steatohepatitis (NASH) (Ascha et al., 2010[8]). HCV does not integrate into its host genome and has a cytoplasmic life cycle (Moradpour et al., 2007[130]). HCC, therefore, must involve several indirect mechanisms including the interplay between HCV and host cell genes/proteins for pathological consequences. In addition, HCV induces epithelial to mesenchymal transition (EMT) state that is known as important element in cancer progression (Bose et al., 2012[24]). This review will discuss recent advances in HCV research with a focus on establishment of chronicity and liver disease progression.
Evasion of innate/ adaptive immune responses by HCV
IFN response
HCV infection is sensed by multiple innate immune pathways, but often not cleared by immune responses, resulting in a chronic infection. HCV blocks the IFN response pathway by several mechanisms. HCV NS3/4A utilizes its protease domain to cleave key innate immune signaling adaptor proteins, effectively inactivating viral RNA detection program (Horner, 2014[74]). HCV NS3/4A protein cleaves MAVS and TRIF (Baril et al., 2009[15]; Li et al., 2005[105]; Lin et al., 2006[108]; Loo et al., 2006[111]; Meylan et al., 2005[126]; Li et al., 2005[104]), and can alter RIG-I and TLR signaling pathway. Hepatocytes persistently infected with HCV and treated with IFN-α, PKR kinase is activated (Kang et al., 2009[85]) for translational suppression of host mRNAs, including ISGs, and antiviral functions of IFN (Garaigota and Chisari, 2009[60]). Several HCV proteins have been implicated as regulators of the IFN response pathway. Expression of HCV proteins blocks IFN signaling at the level of the JAK/STAT pathway (Heim et al., 1999[72]; Raychoudhuri et al., 2011[157]), and impairs IRF-7 nuclear localization through its NS5A protein (Raychoudhuri et al., 2010[158]; Chowdhury et al., 2014[34]).
IFI6 is a type I ISG and plays a critical role in the regulation of apoptosis. IFI6 is strongly associated with the immune system, but its antiviral effects are not well known. Our recent (unpublished) experimental findings suggest co-localization of HCV co-receptors during HCV entry are compromised by IFI6 mediated disruption of kinase function, thereby inhibiting HCV at the point of entry.
Cytokine response
A relationship between the activation of genes involved in the IL-6 signaling pathway and the development of HCC has been observed (Zekri et al., 2009[201]). An increase of the β-2 microglobulin in serum level as well as IL-6 level was observed among HCV infected HCC patients. Weakening of the immune system, due to IL-6, may be responsible for a more severe progression of HCC and the hyperexpression of β-2 microglobulin (Tang et al., 2008[179]). HCV core protein attenuates IL-6 stimulated acute-phase response, and contributes to impaired innate immunity for viral persistence (Malaguarnera et al., 2000[116]; Ait-Goughoulte et al., 2010[2]). TNF-α plays diverse roles, including in the inflammatory processes, in HCV infection (Saito et al., 2006[164]). HCV may actively contribute to the fibrogenic process via the paracrine effect of IL-8 secreted by infected hepatocytes (Koike and Moriya, 2005[98]).
Autophagy
Autophagy is a process of degradation of cytoplasmic materials, including damaged organelles and long-lived proteins, in the cells for the maintenance of cellular homeostasis. During autophagy, the double-membrane vesicles, called autophagosome, engulf the cytoplasmic materials and fuse with the lysosome for degradation. Autophagy has been identified as a component of the innate immune system against viral infection. We were the first to demonstrate that HCV induces autophagy in immortalized human hepatocyte (Ait-Goughoulte et al., 2008[3]). Subsequently, HCV subgenomic replicon and infection were shown to induce autophagy in hepatoma cells (Sir et al., 2008[177]; Dreux et al., 2009[45]; Mizui et al., 2010[127]). Autophagy proteins (Beclin-1, Atg4B, Atg5 and Atg12) are required for initiation of HCV replication (Dreux et al., 2009[45]) and contribute to the effective production of virus particles (Tanida et al., 2009[180]). Recently, we have shown that knockdown of autophagy proteins in HCV infected hepatocytes enhance interferon signaling pathway and induces apoptosis (Shrivastava et al., 2011[176]). HCV mediated autophagy may promote infectious virus particle production and evade innate immune response for establishment of persistent infection (Shrivastava et al., 2011[176]; Shrivastava and Ray, 2014[175]).
Complement
The complement system is one of the vital effectors in the innate immune system for targeting and eliminating infected cells and invading microorganisms, including free virus particles (Mollnes et al., 2002[128]; Gasque, 2004[61]; Kim and Song, 2006[93]). HCV escapes the complement response by regulating complement components. HCV proteins suppress C3/C4 complement expression (Mazumdar et al., 2012[122]; Banerjee et al., 2011[10]), and attenuates membrane attack complex (MAC)-mediated microbicidal activity by suppressing C9 expression (Kim et al., 2013[96]). To avert damage from excessive complement activation and MAC formation, host cells express membrane-bound regulators of complement activation (RCA) proteins, including CD46, CD55 and CD59, to limit these processes (Hourcade et al., 2000[75]; Williams et al., 2003[193]; Pangburn et al., 2008[146]). HCV core protein enhances transcription and surface expression of DAF/CD55 in infected hepatocytes and promotes incorporation onto mature HCV particles (Mazumdar et al., 2013[121]). HCV also incorporates CD59 and protects against complement mediated lysis (Amet et al., 2012[7]; Ejaz et al., 2012[46]). DAF/CD55 expression has been associated with complement dependent cytolysis (CDC), antibody dependent cell cytolysis (ADCC), and NK cell function (Finberg et al., 1992[51]; Bellone et al., 2012[19]; Kim et al., 2014[94]). The strategies adopted by HCV to modulate complement pathways imply a significant advantage for survival of chronically infected hepatocytes, enhancing viral fitness in establishing chronicity and liver disease promotion.
Dendritic cell, NK cell, and T cell functions
HCV can have an inhibitory effect on antigen presenting cells, resulting in reduction of antigen-specific T-cell activation. These effects may contribute to the overall low level of immunogenicity of HCV observed in chronically infected patients (Saito et al., 2008[163]). HCV has an inhibitory role on cathepsin S-mediated major histocompatibility complex (MHC) class II maturation, which may contribute to weak immunogenicity of viral antigens in chronically infected humans (Kim et al., 2012[95]). Further, HCV has been shown to attenuate interferon induced MHC class I expression and decreases CD8+ T cell effector functions (Kang et al., 2014[88]). HCV disables a key receptor ligand (MICA/B) in infected hepatocytes, inhibiting the ability of infected cells to respond to stimuli from NK cells to positively regulate complement synthesis (Kim et al., 2014[94]). Reduced NK cell function may also contribute to the emergence of HCC in chronic liver disease. NK cells induce apoptosis in cells that have either down-regulated MHC class I expression or up-regulated stress-induced ligands (Kim et al., 2014[94]). Broad and potent T cell responses (Neumann-Haefelin and Thimme 2013[136]), and a rapid induction of neutralizing antibody responses help in virus clearance (Osburn, 2014[145]).
Antibody response
In contrast to CD8+T cells, viral escape is likely not a major determinant of HCV specific CD4+ T cell failure (Fleming et al., 2010[52]; Fuller et al., 2010[59]). This is in agreement with the observation that HCV specific CD4+ T cell responses are very weak and dysfunctional in chronic infection and also in agreement with the concept that HCV specific CD4+ T cells primarily have a helper function rather than strong direct antiviral activity. Although, HCV specific neutralizing antibodies exist in infected patients, the virus often escapes from humoral immune response by multiple mechanisms inhibiting evolution of viral quasispecies and display mutations within targeted epitopes (Farci et al., 1996[49]; von Hahn et al., 2007[190]; Dowd et al., 2009[44]; Di Lorenzo et al., 2011[43]). Most neutralizing antibodies show little cross-neutralization of heterologous viral strains; thus, identification of neutralizing antibodies with broad cross-neutralizing activity is an important prerequisite for the use of neutralizing antibodies in prophylactic or therapeutic vaccination strategies. In addition, HCV particles are protected though interaction of envelope glycoproteins with lipoproteins (Mazumdar et al., 2011[120]) and scavenger receptor B1 (Scarselli et al., 2002[166]; Bartosch et al., 2003[17]) in facilitating virus entry into mammalian cells.
The important roles of viral escape in evasion from the neutralizing antibody response have been supported from study performed in patients who underwent liver transplantation (Fafi-Kremer et al., 2010[48]). Reinfection of the liver graft included only few viral quasispecies that were present in the explanted liver. The quasispecies that established reinfection were resistant to homologous neutralizing antibodies, indicating viral escape, while the viral quasispecies that were lost after transplantation were sensible to neutralization by homologous antibodies. A HCV candidate vaccine phase I clinical trial was conducted at the Saint Louis University Vaccine and Treatment Evaluation Unit with the recombinant E1 and E2 glycoproteins in human volunteers, and suggested modest immunogenicity (Frey et al., 2010[54]; Ray et al., 2010[154]; Meyer et al., 2011[125]; Ray, 2011[153]). A different study aimed to elicit HCV specific T cells using a recombinant adenoviral vector strategy in a phase I study in human healthy volunteers (Barnes et al., 2012[16]). The results suggested generation of broad, long-lasting, and functional T cell responses. The protective nature of these responses against HCV exposure remains to be understood.
HCV infection and met metabolic disorders
The metabolic syndrome is a constellation of problems that includes insulin resistance, obesity, hepertension, and hyperlipidemia. Increasingly, components of the metabolic syndrome are being linked to various forms of cancer with respect to both increased risk of disease and worsened outcome. HCV induced insulin resistance impairs antiviral effect of interferon (El-Zayadi and Anis, 2012[47]). Possible explanations for the unique association between insulin resistance and HCV infection may be related to differences in the clinical course of liver inflammation and fibrosis, or in the mode of TNF-receptor activation or cleavage (Joyce et al., 2009[81]). Marked increases in both sTNFR1 and sTNFR2 were demonstrated in HCV-diabetic patients (Shintani et al., 2004[171]). Insulin resistance, a link among chronic HCV infection, TNF-α, and type 2 diabetes (T2D) possibly exists in the correlation with liver disease (Knobler et al., 2003[97]; Sheikh et al., 2008[169]; Ray et al., 1998[155]; Ghosh et al., 2000[63]; Saito et al., 2006[164]; Bose and Ray, 2014[25]).
We have reported that HCV infection upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream Akt/protein kinase B signaling pathway for insulin resistance (Banerjee et al., 2008[13]) via mTOR/S6K1 pathway (Bose et al., 2012[ref:24]). Insulin resistance is paradoxically associated within a reduced ability of insulin signaling to inhibit glucose production, whereas insulin-stimulated lipogenesis is enhanced in the liver and two forkhead transcription factors, FoxO1 and FoxA2 to play important roles in this process. HCV can differentially modulate activation of forkhead transcription factors and insulin induced metabolic gene expression (Banerjee et al., 2010[11]; Bose et al., 2014[25]).
Insulin resistance and subsequent hyperinsulinemia are highly associated with fatty liver disease and are important risk factors for the progression of fibrosis in chronic hepatitis C (Sheikh et al., 2008[169]; Banerjee et al., 2010[12]; Ortiz et al., 2002[144]). Hepatitis C resembles non-alcoholic steatohepatitis (NASH) in numerous features from metabolic aspect, such as the presence of steatosis, serum dyslipidemia, and oxidative stress in the liver (Bugianesi et al., 2004[29]). Steatosis is prevalent with HCV genotype 3 infection and correlates with the level of HCV replication (Adinolfi et al., 2013[1]; Roingeard, 2013[161]). HCV related steatosis predicts an advanced liver disease and a more rapid progression of fibrosis, as well as an increased risk of development of HCC. Viral fatty liver may not impact on response to therapy, while metabolic steatosis does (Negro, 2012[135]). Similarly, viral insulin resistance may not reduce the rate of response to therapy to the same extent that metabolic insulin resistance does.
Micro RNAs
Micro RNAs (miRNAs) constitute a class of ~18-22 nucleotides long non-coding RNAs and play a crucial role in the regulation of gene expression. Deregulation of miRNA occurs frequently in a variety of diseases, including liver (Roberts et al., 2011[160]). The major targets and functions of specific miRNAs vary under different physiological or pathological conditions and in different cell types. Several RNA viruses interact directly with cellular miRNAs in modulating viral genome replication (Cullen, 2013[39]; Conrad and Niepmann, 2014[36]). The liver specific miR-122 is required for efficient HCV replication. HCV binds two molecules of this liver-specific miR-122 resulting in a novel, unprecedented up-regulation of the viral genome (Jopling et al., 2005[80]). Sequestration of miR-122 in HCV-infected cultured cells or in livers of infected chimpanzees leads to a dramatic loss of infectious virus without emergence of resistant virus (Lanford et al., 2010[101]). A phase 2 study using miR-122 antagonist (miravirsen) indicated effective anti-HCV activity (Janssen et al., 2013[79]). Other miRNAs, such as miR-199a*, let-7b, miR-196 and miR-448, physically interact with HCV RNA and suppress the viral replication. HCV infection can alter miRNA expression profile of the host cell in facilitating escape from immune system (Shrivastava et al., 2013[174]; Cullen, 2013[40]). We have previously shown that miR-130a expression is up-regulated in HCV infected cells, and facilitates virus replication by inhibiting interferon-induced transmembrane protein IFITM1 (Bhanja Chowdhury et al., 2012[20]). HCV infection suppresses miR-181c expression by down-regulating transcription factor C/EBP-ß and promotes HOXA1 expression, which subsequently upregulates STAT3 and STAT5 expression (Mukherjee et al., 2014[132]). In addition, exogenous expression of miR-181c restricts HCV replication by binding with E1 and NS5A.
Inflammation, fibrosis/ cirrhosis, hepatocellular carcinoma
Inflammation
An inflammatory process resulting from infection and/or tissue damage is an early defense mechanism during which striking changes in protein synthesis occur mainly in the liver. Inflammatory cells and mediators are found frequently in the local environment of tumors, and inflammation is considered a hallmark of cancer (Hanahan and Weinberg, 2011[66]). Kupffer cells are resident macrophages in the liver and play a pivotal role in triggering inflammation during liver diseases (Zimmermann et al., 2012[205]). HCV infection is sensed by pattern recognition receptors (PRRs) on Kupffer cells and modulates inflammatory responses (Liaskou et al., 2012[107]). Our recent results demonstrated that monocyte-derived human macrophages (THP-1) incubated with cell culture grown HCV enhance the secretion of IL-1β/IL-18 into culture supernatants (Shrivastava et al., 2013[173]). A similar cytokine release was observed from peripheral blood mononuclear cells (PBMCs) derived primary human macrophages and Kupffer cells upon incubation with HCV. Macrophage cell line (THP-1) incubated with HCV led to caspase-1 activation and release of proinflammatory cytokines. HCV induces pro-IL-1β and pro-IL-18 synthesis via the NF-κB signaling pathway in macrophages, although consequence of these proinflammatory cytokine syntheses in liver pathogenesis remains to be elucidated.
Fibrosis
Hepatocellular injury followed by inflammation and activation of the innate immune system may lead to early stage liver fibrosis, resulting in hepatic stellate cell (HSC) activation (Hernandez-Gea and Friedman, 2011[73]). Activated HSCs are both signaling and target cells for a great variety of stimulatory and inhibitory fibrogenic cytokines and growth factors. Hepatic fibrosis affects a large number of people worldwide, and contributes to the processes and pathways involved in malignant transformation. In fibrotic tissues, myofibroblasts accumulate and secrete an excessive amount of collagen that is deposited as fibers, thereby compromising organ function and leading to its failure. Quiescent stellate cells undergo activation to adopt myofibroblast morphology and secrete type I collagen, the principal matrix protein responsible for the development of liver fibrosis, cirrhosis and cancer progression (Kang et al., 2011[86]; Zhang and Friedman, 2012[202]).
Stellate cells produce growth factors, including interleukin 6, hepatocyte growth factor, and Wnt ligands, fostering an environment for hepatocyte proliferation (Friedman, 2008[55], [56]). Similarly, hepatic myofibroblasts can enhance growth and migration of malignant hepatocytes, at least partially through platelet-derived growth factor (PDGF) and transforming growth factor-β (TGF-β) mediated mechanisms (van Zijl et al., 2009[188]). TGF-β signaling are highly dependent on extracellular matrix (ECM) interactions. TGF-β is directly recruited to the ECM by latent TGF-β binding protein (LTBPs), which have affinity for both TGF-β and ECM fibrils. When bound to LTBP, TGF-β are unable to signal. This suggests that accumulation of ECM would lead to increased proliferation and decreased apoptosis, because TGF-β signaling would be suppressed. However, LTBPs contain multiple proteinase sensitive sites, and cleavage of those sites by MMPs leads to the release of TGF-β (Todorovic and Rifkin, 2012[186]). In the setting of inflammation or increased migratory potential, elevated MMP activity can liberate sequestered TGF-β. Fibrotic ECM, containing more sequestered TGF-β, would release greater amounts of the cytokine. This could antagonize oncogenesis by inhibiting proliferation and promoting apoptosis. HCV core upregulates the expression of TGF-β (Torre et al., 1994[187]; Taniguchi et al., 2004[181]), and NS5A modulates TGF-β signaling through interaction with TGF-β receptor I (Shin et al., 2005[171]). Another study showed that different thresholds of Smad3 activation control TGF-ß responses in hepatocytes and that liver cancer-derived HCV core protein, by decreasing Smad3 activation, switches TGF-β growth inhibitory effects to tumor-promoting responses (Matsuzaki et al., 2007[119]). HCV core also triggers the production of both TGF-β2 and VEGF proteins through multiple pathways (Battaglia et al., 2009[18]). As HCV infected livers progress from chronic hepatitis to cirrhosis and/or HCC, pSmad3L/PAI-1 increases with fibrotic stage and necroinflammatory grade; pSmad3C/p21 decreases (Choi and Hwang, 2006[32]). HCV infected hepatocytes release TGF-β1 and other profibrogenic factors that differentially modulate expression of several key genes that can activate HSCs in liver fibrosis (Schulze-Krebs et al., 2005[167]). CD81 protein, a key entry coreceptor for HCV, is highly expressed in HSCs (Mazzocca et al., 2002[123]) and HCV E2 protein can directly interact with CD81 on HSC surface, inducing fibrogenic effects on HSCs (Mazzocca et al., 2005[124]). Therefore, it is possible that chronic inflammation associated with HCV infection shifts hepatocytic TGF-β signaling from tumor suppression to fibrogenesis, accelerating liver fibrosis and increasing the risk of HCC.
Fibrosis is defined by changing the amount and composition of ECM component, which contribute to tumorigenesis. Integrin family of transmembrane receptors contributes to increase deposition of fibrillar collagen type I and III, as well as fibronectin in hepatic fibrosis. In addition to the fibrillar collagens, other ECM molecules including laminin, fibronectin, and several nonfibrillar collagens may also amplify carcinogenic signaling. Although these proteins are in relatively low abundance compared to the fibrillar collagens, their potential function as growth factor receptor ligands could amplify their carcinogenic impact. Increased ECM may stimulate integrin signaling in hepatocyte. Integrins promote growth and survival by activating phosphoinositide 3 kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling cascade (Cox et al., 2010[38]), thereby enhancing the growth and survival of precancerous cells. The correlation of collagen expression, integrin expression and tumorigenicity is studied in human and animal HCC specimens (Lee et al., 2009[102]; Lai et al., 2011[99]). Other mechanisms for integrin-mediated tumorigenesis are increased migration (Fransvea et al., 2009[53]; Fu et al., 2010[57]) and survival through anti-apoptotic signaling (Zhang et al., 2002[203]). In tumor cell lines, overexpression of integrin B1 actually leads to growth arrest, attributed to up-regulation of the cyclin-dependent kinase inhibitor p21 and p27. In addition, human HCC samples have decreased expression of integrin B3, and its overexpression in a human HCC cell line leads to apoptosis (Wu et al., 2009[195]).
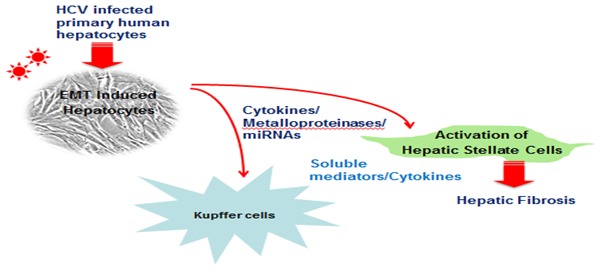
Interestingly, cell tracing studies have shown that a significant portion of these myofibroblasts arise from the conversion of epithelial cells through an EMT process (Iwano et al., 2002[78]). Hepatocytes can undergo EMT and contribute significantly to liver fibrosis (Figure 1(Fig. 1)). Indeed, lineage-tracing in transgenic mice also indicates that hepatocytes undergo EMT during CCl4 induced liver fibrosis (Zeisberg et al., 2007[200]). Interestingly, hepatocytes derived from cirrhotic livers also display characteristics of EMT, which has implications for the progression to HCC (Nitta et al., 2008[138]). We have shown that primary human hepatocytes infected in vitro with cell culture grown HCV display morphological and molecular alterations suggestive of EMT, and display an extended life span (Bose et al., 2012[24]). Similar observations have been noted in continuous cell types (Akkari et al., 2012[4]; Wilson et al., 2012[194]; Conti et al., 2013[37]; Iqbal et al., 2014[77]). EMT type II has been linked to escape from senescence and apoptosis, which suggest a role in epithelial cell growth promotion. Among HCV proteins, core and NS5A are suggested to induce EMT (Akkari et al., 2012[4]; Quan et al., 2014[152]). EMT is likely to play a major mechanism in tumor progression, local invasion, metastasis, and therapeutic resistance; and is linked to the development of stem-like properties by cancer cells (Mani et al., 2008[117]; Thiery et al., 2009[185]).
Figure 1. Pathways associated with fibrogenic potential of hepatic stellate cells.
Hepatocellular carcinoma
Activated HSCs and myofibroblasts may directly support hepatic tumorigenesis and invasion of primary tumors (Kalluri and Zeisberg, 2006[83]). Desmoplasia or cancer associated fibrosis is the growth of fibrous or connective tissue that usually occurs around a malignant neoplasm, causing dense fibrosis around the tumor (Kang et al., 2011[86]; Zhang and Friedman, 2012[202]; Yaqoob et al., 2012[196]; Liu et al., 2013[109]). Several studies have identified cells resembling activated stellate cells associated with the liver progenitor cell niche, suggesting that these cells may provide paracrine signals that promote stem cell expansion (Greenbaum and Wells, 2011[65]). The nature of these paracrine signals, and the mechanisms underlying the supportive role of HSCs in stem cell expansion, are currently unknown and of intense interest. Intercellular signaling networks exist between tumors and tumor-associated fibroblasts. Tumor secretion of PDGF and TGF-β causes to changes in ECM composition and organization through stimulating myofibroblast activation. In addition, hepatic stellate cells secrete more angiopoietin 1 when activated (Taura et al., 2008[183]), facilitating an angiogenic milieu that is supportive of tumor growth.
Tumors may signal to surrounding stroma. For example, elevated hedgehog signaling has been associated with liver injury in mice and humans (Jung et al., 2008[82]; Lees et al., 2005[103]), and promotes liver regeneration (Ochoa et al., 2010[142]). Hedgehog signaling from tumors to the stromal microenvironment may be responsible for promoting tumor progression (Yauch et al., 2008[198]). Since hedgehog signaling may induce EMT, the tumorigenic effect of hedgehog could be mediated by increased myofibroblast activation and fibrosis (Omenetti et al., 2008[143]; Syn et al., 2009[178]; Philips et al., 2011[150]).
Increased stromal stiffness precedes and accompanies fibrosis in chronic liver disease (Georges et al., 2007[62]; Yin et al., 2007[199]), and elevated liver stiffness is associated with enhanced risk of HCC (Masuzaki et al., 2008[118]). Stromal stiffness increases activation of stellate cells (Wells, 2008[192]) and portal fibroblasts (Li et al., 2007[106]), creating a positive feedback loop that continues to promote fibrosis. Stromal stiffness is regulated in part by matrix metalloproteinases (MMPs) and their inhibitors, but MMPs can regulate cell proliferation independently of their effects on stromal stiffness. Although MMPs degrade the stroma, they paradoxically increase HSC proliferation, liver growth, and tumor progression (Theret et al., 2001[184]; Nishio et al., 2003[137]; Zhou et al., 2006[204]).
HCC diagnosed in cirrhotic and non-cirrhotic livers may display different imaging and pathological attributes such as size, differentiation, and encapsulation (Brancatelli et al., 2002[27]). When associated with NAFLD, HCC is often moderately or well differentiated and occurs as solitary large mass (Regimbeau et al., 2004[159]; Bugianesi et al., 2002[28]). HCC with mild or no fibrosis may share these characteristics (Yasui et al., 2011[197]; Kawada et al., 2009[92]; Iannaccone et al., 2007[76]). Similarly, HCC complicating the metabolic syndrome and arising in non-fibrotic livers often remains well differentiated despite a larger size (Paradis et al., 2009[147]). Deregulation of the Wnt/β-catenin pathway has little role in the development of HCC associated with the metabolic syndrome in the absence of significant liver fibrosis (Paradis et al., 2009[147]).
Alcoholic liver disease (ALD) is the most common cause of HCC, accounting for approximately one-third of all HCC cases (Morgan et al., 2004[131]). Alcohol abuse has synergistic effects with other risk factors for the development of HCC, such as infection with HBV or HCV, diabetes and obesity (Hassan et al., 2002[68]; Loomba et al., 2010[112]).
Oncogenic potential of HCV proteins
Highly conserved HCV core protein is related to the induction of liver steatosis in transgenic mice and in HCV infected patients (Moradpour et al., 1996[129]; Rouille et al., 2006[162]; Barba et al., 1997[14]). HCV core protein is involved in apoptotic changes, glucose and lipid metabolism, and malignant transformation. Among many interactions with cellular factors, HCV core has been shown to induce ROS production via interaction with heat shock protein Hsp60 (Kang et al., 2009[87]), and modulates expression of the tumor suppressor protein p53 (Ray et al., 1997[156]; Lu et al., 1999[113]), p73 (Alisi et al., 2003[6]) and pRb (Cho et al., 2001[31]). Core also inhibits the expression of the cyclin-dependent kinase (CDK) inhibitor p21/Waf (Wang et al., 2002[191]). p21 is a transcriptional target of p53 and blocks the cyclin/CDK complexes involved in cell cycle control and tumor formation. Core induces activation of the Raf1/MAPK pathway (Hayashi et al., 2000[69]), protects cells from serum starvation and growth arrest and drives cells into proliferation. HCV core also activates the Wnt/β-catenin pathway, which controls cell proliferation, DNA synthesis and cell-cycle progression (Fukutomi et al., 2005[58]). We have shown that HCV core protein acts as a positive regulator in AR signaling, providing further insight into oncogenic potential in the development of HCC in HCV infected individuals (Kanda et al., 2008[84]). HCV core protein behaves as a positive regulator in androgen receptor signaling and enhances the expression of VEGF in hepatocytes (Hassan et al., 2009[67]). HCV NS2 also retains p53 into the cytoplasm, although the mechanism is not well understood (Bittar et al., 2013[21]). A direct role of HCV NS3 was reported in the neoplastic transformation of hepatocytes in vivo and in vitro (Sakamuro et al., 1995[165]). The NS3 protein also forms complexes with p53, and inhibits p21 promoter activity (He et al., 2003[70]). HCV NS5A protein interacts with various signaling pathways including cell cycle/apoptosis (Kasprzak and Adamek, 2008[90]) in host cells and shares some signaling targets with core protein. NS5A is recognized as a transcriptional activator for many target genes (Kato et al., 1997[91]). Transcription factor IID activities are modified by NS5A in the suppression of p53-dependent transcriptional transactivation and apoptosis (Lan et al., 2002[100]; Majumder et al., 2001[115]). NS5A also interacts with pathways, such as Bcl2 (Chung et al., 2003[35]), phosphatidylinositol 3-kinase (PI3-K) (He et al., 2002[71]), Wnt/beta catenin signaling (Park et al., 2009[148]), and mTOR (Peng et al., 2010[149]) to activate cell proliferation signaling and inhibits apoptosis. HCV polymerase NS5B forms a cytoplasmic complex with Rb in infected cells (Munakata et al., 2007[133]). NS5B dependent down-regulation of Rb leads to activation of E2F-dependent transcription and increases cell proliferation. The interaction of the NS5B with Rb results in the degradation of Rb and activates the MAD2 promoter (Munakata et al., 2005[134]). Thus, infection with HCV may lead to a loss of host-cell genomic stability due to deregulation of Rb pathway. The integrity of Rb appears to be important in the normally quiescent hepatocytes, as liver-specific loss of Rb may promote ectopic cell-cycle entry, aberrant ploidy and neoplastic transformation (Machida et al., 2009[114]).
Conclusions
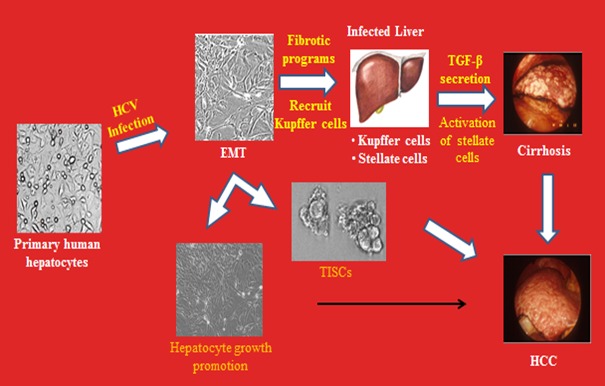
HCV remains a major cause of cirrhosis, liver failure and HCC despite recent dramatic advances in antiviral treatment. Some patients may experience progression of liver disease or HCC despite viral clearance. Trace amounts of HCV RNA from successfully treated patients can be infectious (Veerapu et al., 2014[189]). We do not know the long-term efficacy of treatment with the new generation of DAAs, particularly with interferon-free regimens; and generation of potential resistant virus (Di Bisceglie et al., 2014[42]; Poveda et al., 2014[151]). Thus, it is important to understand the underlying mechanisms of interferon mediated and interferon free DAA mediated clearance of chronic HCV infection. Safety profile of DAAs with side effects, especially in patients with advanced liver fibrosis is also an important point for consideration (D’Ambrosio and Colombo, 2013[41]). The treatment may not work well upon repeated reinfection, especially among the drug addicts. An effective vaccine against multiple genotypes along with DAAs will be most appropriate to combat HCV infection. While there is evidence of a strong link between chronic HCV infection, fibrosis/cirrhosis, and HCC, how HCV promotes the disease processes is under intense investigation. HCV causes persistent infection, although the viral genome does not integrate into the host cell genome. Somatic cells have the ability to become pluripotent cells when transiently exposed to strong stimuli that they would not normally experience in their living environments (Obokata et al., 2014[141]). This reprogramming does not require nuclear transfer or genetic manipulation (Obokata et al., 2014[140]). Primary human hepatocytes, when infected in vitro with cell culture grown HCV, display an extended life span, and morphological and molecular alterations suggestive of epithelial-mesenchymal transition (EMT) state and tumor initiating stem cell (TISC) generation (Figure 2(Fig. 2)). This may promote to fibrosis/cirrhosis and HCC, and needs investigations to unveil the underlying mechanisms and overlaps in developing appropriate therapeutic modalities.
Figure 2. Hypothesis for HCV induced fibrosis/cirrhosis and HCC in chronically infected liver.
Acknowledgements
We thank Lin Cowick for preparation of the manuscript. The current research of R.B.R. is supported by the Internal Liver Center Fund of Saint Louis University and grant R01DK081817 and research of R.R. is supported by the Internal Blue Ribbon Fund and Presidential Research Funds of Saint Louis University, grants R01DK080812 and U54AI057160 for the Midwest Center for Excellence from the National Institutes of Health. We feel sorry for not citing the work of many other HCV investigators owing to space constraints.
References
- 1.Adinolfi LE, Restivo L, Marrone A. The predictive value of steatosis in hepatitis C virus infection. Expert Rev Gastroenterol Hepatol. 2013;7:205–213. doi: 10.1586/egh.13.7. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Goughoulte M, Banerjee A, Meyer K, Mazumdar B, Saito K, Ray RB, et al. Hepatitis C virus core protein interacts with fibrinogen-beta and attenuates cytokines stimulated acute-phase response. Hepatology. 2010;51:1505–1513. doi: 10.1002/hep.23502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akkari L, Gregoire D, Floc’h N, Moreau M, Hernandez C, Simonin Y, et al. Hepatitis C viral protein NS5A induces EMT and participates in oncogenic transformation of primary hepatocyte precursors. J Hepatol. 2012;57:1021–1028. doi: 10.1016/j.jhep.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 5.Ali N, Allam H, May R, Sureban SM, Bronze MS, Bader T, et al. Hepatitis C virus-induced cancer stem cell-like signatrues in cell culture and murine tumor xenografts. J Virol. 2011;85:12292–12303. doi: 10.1128/JVI.05920-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alisi A, Giambartolomei S, Cupelli F, Merlo P, Fontemaggi G, Spaziani A, et al. Physical and functional interaction between HCV core protein and the different p73 isoforms. Oncogene. 2003;22:2573–2580. doi: 10.1038/sj.onc.1206333. [DOI] [PubMed] [Google Scholar]
- 7.Amet T, Ghabril M, Chalasani N, Byrd D, Hu N, Grantham A, et al. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology. 2012;55:354–363. doi: 10.1002/hep.24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology. 2010;51:1972–1978. doi: 10.1002/hep.23527. [DOI] [PubMed] [Google Scholar]
- 9.Au JS, Pockros PJ. Novel therapeutic approaches for hepatitis C. Clin Pharmacol Ther. 2014;95:78–88. doi: 10.1038/clpt.2013.206. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee A, Mazumdar B, Meyer K, Di Bisceglie AM, Ray RB, Ray R. Transcriptional repression of C4 complement by hepatitis C virus proteins. J Virol. 2011;85:4157–4166. doi: 10.1128/JVI.02449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee A, Meyer K, Mazumdar B, Ray RB, Ray R. Hepatitis C virus differentially modulates activation of forkhead transcription factors and insulin-induced metabolic gene expression. J Virol. 2010;84:5936–5946. doi: 10.1128/JVI.02344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee A, Ray RB, Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108–2133. doi: 10.3390/v2092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee S, Saito K, Ait-Goughoulte M, Meyer K, Ray RB, Ray R. Hepatitis C virus core protein upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream akt/protein kinase B signaling pathway for insulin resistance. J Virol. 2008;82:2606–2612. doi: 10.1128/JVI.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med. 2012;4:115ra1. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 18.Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL, et al. Liver cancer-derived hepatitis C virus core proteins shift TGF-beta responses from tumor suppression to epithelial mesenchymal transition. PLoS One. 2009;4:e4355. doi: 10.1371/journal.pone.0004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellone S, Roque D, Cocco E, Gasparrini S, Bortolomai I, Buza N, et al. Downregulation of membrane complement inhibitors CD55 and CD59 by siRNA sensitizes uterine serous carcinoma overexpressing Her2/neu to complement and antibody-dependent cell cytotoxicity in vitro : implications for trastuzumab-based immunotherapy. Br J Canc. 2012;106:1543–1550. doi: 10.1038/bjc.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol. 2012;86:10221–10225. doi: 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittar C, Shrivastava S, Bhanja Chowdhury J, Rahal P, Ray RB. Hepatitis C virus NS2 protein inhibits DNA damage pathway by sequestering p53 to the cytoplasm. PLoS One. 2013;8:e62581. doi: 10.1371/journal.pone.0062581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Bose SK, Kim H, Meyer K, Wolin N, Davidson NO, Ray R. Forkhead box transcription factor regulation and lipid accumulation by hepatitis C virus. J Virol. 2014;88:4195–4203. doi: 10.1128/JVI.03327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose SK, Meyer K, Di Bisceglie AM, Ray RB, Ray R. Hepatitis C virus induces epithelial-mesenchymal transition in primary human hepatocytes. J Virol. 2012;86:13621–13628. doi: 10.1128/JVI.02016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bose SK, Ray R. Hepatitis C virus infection and insulin resistance. World J Diabetes. 2014;5:52–58. doi: 10.4239/wjd.v5.i1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bose SK, Shrivastava S, Meyer K, Ray RB, Ray R. Hepatitis C virus activates the mTOR/S6K1 signaling pathway in inhibiting IRS-1 function for insulin resistance. J Virol. 2012;86:6315–6322. doi: 10.1128/JVI.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brancatelli G, Federle MP, Grazioli L, Carr BI. Hepatocellular carcinoma in noncirrhotic liver: CT, clinical, and pathologic findings in 39 U.S. residents. Radiology. 2002;222:89–94. doi: 10.1148/radiol.2221010767. [DOI] [PubMed] [Google Scholar]
- 28.Bugianesi E, Leone N, Vanni E, Marchesini G, Brunello F, Carucci P, et al. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123:134–140. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- 29.Bugiansesi E, Manzini P, D’Antico S, Vanni E, Longo F, Leone N, et al. Relative contribution of iron burden, HFE mutations, and insulin resistance to fibrosis in nonalcoholic fatty liver. Hepatology. 2004;39:179–187. doi: 10.1002/hep.20023. [DOI] [PubMed] [Google Scholar]
- 30.Casey LC, Lee WM. Hepatitis C virus therapy update. 2013. Curr Opin Gastroenterol. 2013;29:243–249. doi: 10.1097/MOG.0b013e32835ff972. [DOI] [PubMed] [Google Scholar]
- 31.Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb down-regulation and E2F-1 up-regulation. Biochem Biophys Acta. 2001;1538:59–66. doi: 10.1016/s0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- 32.Choi SH, Hwang SB. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J Biol Chem. 2006;281:7468–7478. doi: 10.1074/jbc.M512438200. [DOI] [PubMed] [Google Scholar]
- 33.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 34.Chowdhury JB, Kim H, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates IRF-7-mediated interferon-α signaling. J Interferon Cytokine Res. 2014;34:16–21. doi: 10.1089/jir.2013.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung YL, Sheu ML, Yen SH. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int J Cancer. 2003;107:65–73. doi: 10.1002/ijc.11303. [DOI] [PubMed] [Google Scholar]
- 36.Conrad KD, Niepmann M. The role of microRNAs in hepatitis C virus RNA replication. Arch Virol. 2014;159:849–862. doi: 10.1007/s00705-013-1883-4. [DOI] [PubMed] [Google Scholar]
- 37.Conti B, Minutolo A, Arciello M, Balsano C. Are Hedgehog and Wnt/β-catenin pathways involved in hepatitis C virus-mediated EMT? J Hepatol. 2013;58:636–637. doi: 10.1016/j.jhep.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 38.Cox D, Brennan M, Moran N. Integrins as therapeutic targets: lessons and opportunities. Nat Rev Drug Discov. 2010;9:804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 39.Cullen BR. How do viruses avoid inhibition by endogenous cellular microRNAs? PLoS Pathog. 2013;9:e1003694. doi: 10.1371/journal.ppat.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14:205–210. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Ambrosio R, Colombo M. Safety of direct antiviral agents in real life. Dig Liver Dis. 2013;45(Suppl 5):S363–S366. doi: 10.1016/j.dld.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Di Bisceglie AM, Janczweska-Kazek E, Habersetzer F, Mazur W, Stanciu C, Carreno V, et al. Efficacy of immunotherapy with TG4040, peg-interferon, and ribavirin in a phase 2 study of patients with chronic HCV infection. Gastroenterology. 2014;147:119–131. doi: 10.1053/j.gastro.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Di Lorenzo C, Angus AG, Patel AH. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses. 2011;3:2280–2300. doi: 10.3390/v3112280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology. 2009;136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ejaz A, Steinmann E, Banki Z, Anggakusuma, Khalid S, Lengauer S, et al. Specific acquisition of functional C59 but not CD46 or CD55 by hepatitis C virus. PLOS One. 2012;7:e45770. doi: 10.1371/journal.pone.0045770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Zayadi AR, Anis M. Hepatitis C virus induced insulin resistance impairs response to antiviral therapy. World J Gastroenterol. 2012;18:212–224. doi: 10.3748/wjg.v18.i3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fafi-Kremer S, Fofana I, Soulier E, Carolla P, Meuleman P, Leroux-Roels G, et al. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J Exp Med. 2010;207:2019–2031. doi: 10.1084/jem.20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum againt the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;24:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 51.Finberg RW, White W, Nicholson-Weller A. Decay-accelerating factor expression on either effector or target cells inhibits cytotoxicity by human natural killer cells. J Immunol. 1992;149:2055–2060. [PubMed] [Google Scholar]
- 52.Fleming VM, Harcourt G, Barnes E, Klenerman P. Virological footprint of CD4+ T-cell responses during chronic hepatitis C virus infection. J Gen Virol. 2010;91:1396–1406. doi: 10.1099/vir.0.017699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fransvea E, Mazzocca A, Antonaci S, Giannelli G. Targeting transforming growth factor (TGF)-betaRI inhibits activation of beta1 integrin and blocks vascular invasion in hepatocellular carcinoma. Hepatology. 2009;49:839–850. doi: 10.1002/hep.22731. [DOI] [PubMed] [Google Scholar]
- 54.Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, et al. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine. 2010;28:6367–6373. doi: 10.1016/j.vaccine.2010.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu BH, Wu ZZ, Qin J. Effects of integrins on laminin chemotaxis by hepatocellular carcinoma cells. Mol Biol Rep. 2010;37:1665–1670. doi: 10.1007/s11033-009-9790-1. [DOI] [PubMed] [Google Scholar]
- 58.Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth : correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- 59.Fuller MJ, Shoukry NH, Gushima T, Bowen DG, Callendret B, Campbell KJ, et al. Selection-driven immune escape is not a significant factor in the failure of CD4 T cell responses in persistent hepatitis C virus infection. Hepatology. 2010;51:378–387. doi: 10.1002/hep.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol. 2004;41:1089–1098. doi: 10.1016/j.molimm.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, et al. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–G1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 63.Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173–178. doi: 10.1016/s0168-1702(00)00141-6. [DOI] [PubMed] [Google Scholar]
- 64.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenbaum LE, Wells RG. The role of stem cells in liver repair and fibrosis. Int J Biochem Cell Biol. 2011;43:222–229. doi: 10.1016/j.biocel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 67.Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology. 2009;49:1469–1482. doi: 10.1002/hep.22849. [DOI] [PubMed] [Google Scholar]
- 68.Hassan MM, Hwang LY, Hatten CJ, Swaim M, Li D, Abbruzzese JL, et al. Risk factors for hepatocellular carcinoma : synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206–1213. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- 69.Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology. 2000;32:958–961. doi: 10.1053/jhep.2000.19343. [DOI] [PubMed] [Google Scholar]
- 70.He QQ, Cheng RX, Sun Y, Feng DY, Chen ZC, Zheng H. Hepatocyte transformation and tumor development induced by hepatitis C virus NS3 c-terminal deleted protein. World J Gastroenterol. 2003;9:474–478. doi: 10.3748/wjg.v9.i3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He Y, Nakao H, Tan SL, Polyak SJ, Neddermann P, Vijaysri S, et al. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosyphatidylinositol 3-kinase. J Virol. 2002;76:9207–9217. doi: 10.1128/JVI.76.18.9207-9217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 74.Horner SM. Activation and evasion of antiviral innate immunity by hepatitis C virus. J Mol Biol. 2014;426:1198–1209. doi: 10.1016/j.jmb.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hourcade D, Liszewski MK, Krych-Goldberg M, Atkinson JP. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology. 2000;49:103–116. doi: 10.1016/s0162-3109(00)80296-9. [DOI] [PubMed] [Google Scholar]
- 76.Iannaccone R, Piacentini F, Murakami T, Paradis V, Belghiti J, Hori M, et al. Hepatocellular carcinoma in patients with nonalcoholice fatty liver disease : helical CT and MR imaging findings with clinical pathologic comparison. Radiology. 2007;243:422–430. doi: 10.1148/radiol.2432051244. [DOI] [PubMed] [Google Scholar]
- 77.Iqbal J, McRae S, Mai T, Banaudha K, Sarkar-Dutta M, Waris G. Role of hepatitis C virus induced osteopontin in epithelial to mesenchymal transition, migration and invasion of hepatocytes. PLoS One. 2014;9:e87464. doi: 10.1371/journal.pone.0087464. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 80.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 81.Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, et al. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLOS Pathog. 2009;5:e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung Y, Brown KD, Witek RP, Omenetti A, Yang L, Vandongen M, et al. Accumulation of hedgehog-responsive progenitors parallels alcoholic liver disease severity in mice and humans. Gastroenterology. 2008;134:1532–1543. doi: 10.1053/j.gastro.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 84.Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus core protein augments androgen receptor-mediated signaling. J Virol. 2008;82:11066–11072. doi: 10.1128/JVI.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, et al. PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res. 2009;142:51–56. doi: 10.1016/j.virusres.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 86.Kang N, Gores GJ, Shah VH. Hepatic stellate cells: partners in crime for liver metastases? Hepatology. 2011;54:707–713. doi: 10.1002/hep.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang SM, Kim SJ, Kim JH, Lee W, Kim GW, Lee KH, et al. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-alpha-mediated apoptosis. Cancer Lett. 2009;279:230–237. doi: 10.1016/j.canlet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 88.Kang W, Sung PS, Park SH, Yoon S, Chang DY, Kim S, et al. Hepatitis C virus attenuates interferon-induced major histocompatibility complex class I expression and decreases CD8+T cell effector functions. Gastroenterology. 2014;146:1351–1360. doi: 10.1053/j.gastro.2014.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanwal F, Hoang T, Kramer JR, Asch SM, Goetz MB, Zeringue A, et al. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C virus infection. Gastroenterology. 2011;140:1182–1188. doi: 10.1053/j.gastro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kasprzak A, Adamek A. Role of hepatitis C virus proteins (C, NS3, NS5A) in hepatic oncogenesis. Hepatol Res. 2008;38:1–26. doi: 10.1111/j.1872-034X.2007.00261.x. [DOI] [PubMed] [Google Scholar]
- 91.Kato N, Lan KH, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawada N, Imanaka K, Kawaguchi T, Tamai C, Ishihara R, Matsunaga T, et al. Hepatocellular carcinoma arising from non-cirrhotic nonalcoholic steatohepatitis. J Gastroenterol. 2009;44:1190–1194. doi: 10.1007/s00535-009-0112-0. [DOI] [PubMed] [Google Scholar]
- 93.Kim DD, Song WC. Membrane complement regulatory proteins. Clin Immunol. 2006;118:127–136. doi: 10.1016/j.clim.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 94.Kim H, Bose SK, Meyer K, Ray R. Hepatitis C virus impairs natural killer cell-mediated augmentation of complement synthesis. J Virol. 2014;88:2564–2571. doi: 10.1128/JVI.02988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim H, Mazumdar B, Bose SK, Meyer K, Di Bisceglie AM, Hoft DF, et al. Hepatitis C virus-mediated inhibition of cathepsin S increases invariant-chain expression on hepatocyte surface. J Virol. 2012;86:9919–9928. doi: 10.1128/JVI.00388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim H, Meyer K, Di Bisceglie AM, Ray R. Hepatitis C virus suppresses C9 complement synthesis and impairs membrane attack complex function. J Virol. 2013;87:5858–5867. doi: 10.1128/JVI.00174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knobler H, Zhornicky T, Sandler A, Haran N, Ashur Y, Schattner A. Tumor necrosis factor-alpha-induced insulin resistance may mediate the hepatitis C virus-diabetes association. Am J Gastroenterol. 2003;98:2751–2756. doi: 10.1111/j.1572-0241.2003.08728.x. [DOI] [PubMed] [Google Scholar]
- 98.Koike K, Moriya K. Metabolic aspects of hepatitis C viral infection: steatohepatitis resembling but distinct from NASH. J Gastroenterol. 2005;49:329–336. doi: 10.1007/s00535-005-1586-z. [DOI] [PubMed] [Google Scholar]
- 99.Lai KK, Shang S, Lohia N, Booth GC, Masse DJ, Fausto N, et al. Extracellular matrix dynamics in hepatocarcinogenesis: a comparative proteomics study of PDGFC transgenic and Pten null mouse models. PLoS Genet. 2011;7:e1002147. doi: 10.1371/journal.pgen.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- 101.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee SK, Kim MH, Cheong JY, Cho SW, Yang SJ, Kwack K. Integrin alpha V polymorphisms and haplotypes in a Korean population are associated with susceptibility to chronic hepatitis and hepatocellular carcinoma. Liver Int. 2009;29:187–195. doi: 10.1111/j.1478-3231.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 103.Lees C, Howie S, Sartor RB, Satsangi J. The hedgehog signaling pathway in the gastrointestinal track : implications for development, homeostasis, and disease. Gastroenterology. 2005;129:1696–1710. doi: 10.1053/j.gastro.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 104.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 107.Liaskou E, Wilson DV, Oo YH. Innate immune cells in liver inflammation. Mediators Inflamm. 2012;2012:949157. doi: 10.1155/2012/949157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, et al. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol. 2006;80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu C, Billadeau DD, Abdelhakim H, Leof E, Kaibuchi K, Bernabeu C, et al. IQGAP1 suppresses TβRII-mediated myofibroblastic activation and metastatic growth in liver. J Clin Invest. 2013;123:1138–1156. doi: 10.1172/JCI63836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lok AS, Seeff LB, Morgan TR, di Bisceglie AM, Sterling RK, Curto TM, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136:138–148. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Loomba R, Yang HI, Su J, Brenner D, Iloeje U, Chen CJ. Obesity and alcohol synergize to increase the risk of incident hepatocellular carcinoma in men. Clin Gastroenterol Hepatol. 2010;8:891–898. doi: 10.1016/j.cgh.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 113.Lu W, Lo SY, Chen M, Wu Kj, Fung YK, Ou JH. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology. 1999;264:134–141. doi: 10.1006/viro.1999.9979. [DOI] [PubMed] [Google Scholar]
- 114.Machida K, Liu JC, McNamara G, Levine A, Duan L, Lai MM. Hepatitis C virus causes uncoupling of mitotic checkpoint and chromosomal polyploidy through the Rb pathway. J Virol. 2009;83:12590–12600. doi: 10.1128/JVI.02643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401–1407. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Malaguarnera M, Di Fazio I, Ferlito L, Pistone G, Laurino A, Vinci E, et al. Increase of serum beta2-microglobulin in patients affected by HCV correlated hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2000;12:937–939. doi: 10.1097/00042737-200012080-00014. [DOI] [PubMed] [Google Scholar]
- 117.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou Ay, et al. The epithelial-messenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Masuzaki R, Tateishi R, Yoshida H, Yoshida H, Sato S, Kato N, et al. Risk assessment of hepatocellular carcinoma in chronic hepatitis C patients by transient elastography. J Clin Gastroenterol. 2008;42:839–843. doi: 10.1097/mcg.0b013e318050074f. [DOI] [PubMed] [Google Scholar]
- 119.Matsuzaki K, Murata M, Yoshida K, Sekimoto G, Uemura Y, Sakaida N, et al. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma. Hepatology. 2007;46:48–57. doi: 10.1002/hep.21672. [DOI] [PubMed] [Google Scholar]
- 120.Mazumdar B, Banerjee A, Meyer K, Ray R. Hepatitis C virus E1 envelope glycoprotein interacts with apo-lipoproteins in facilitating entry into hepatocytes. Hepatology. 2011;54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mazumdar B, Kim H, Meyer K, Bose SK, Di Bisceglie AM, Ray RB, et al. Hepatitis C virus infection upregulates CD55 expression on the hepatocyte surface and promotes association with virus particles. J Virol. 2013;87:7902–7910. doi: 10.1128/JVI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mazumdar B, Kim H, Meyer K, Bose SK, Di Bisceglie AM, Ray RB, et al. Hepatitis C virus proteins inhibit C3 complement production. J Virol. 2012;86:2221–2228. doi: 10.1128/JVI.06577-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mazzocca A, Carloni V, Sciammetta S, Cordella C, Pantaleo P, Caldini A, et al. Expression of transmembrane 4 superfamily (TM4SF) proteins and their role in hepatic stellate cell motility and wound healing migration. J Hepatol. 2002;37:322–330. doi: 10.1016/s0168-8278(02)00175-7. [DOI] [PubMed] [Google Scholar]
- 124.Mazzocca A, Sciammetta SC, Carloni V, Cosmi L, Annunziato F, Harada T, et al. Binding of hepatitis C virus envelope protein E2 to CD81 up-regulates matrix metalloproteinase-2 in human hepatic stellate cells. J Biol Chem. 2005;280:11329–11339. doi: 10.1074/jbc.M410161200. [DOI] [PubMed] [Google Scholar]
- 125.Meyer K, Banerjee A, Frey SE, Belshe RB, Ray R. A weak neutralizing antibody response to hepatitis C virus envelope glycoprotein enhances virus infection. PLOS One. 2011;6:e23699. doi: 10.1371/journal.pone.0023699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 127.Mizui T, Yamashina S, Tanida I, Takei Y, Ueno T, Sakamoto N, et al. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J Gastroenterol. 2010;45:195–203. doi: 10.1007/s00535-009-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mollnes TE, Song WC, Lambris JD. Complement in inflammatory tissue damage and disease. Trends Immunol. 2002;23:61–64. doi: 10.1016/s1471-4906(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 129.Moradpour D, Englert C, Wakita T, Wands JR. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- 130.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 131.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 132.Mukherjee A, Shrivastava S, Bhanja Chowdhury J, Ray R, Ray RB. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol. 2014;88:7929–7940. doi: 10.1128/JVI.00787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, et al. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2005;102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Negro F. Steatosis and insulin resistance in response to treatment of chronic hepatitis C. J Viral Hepat. 2012;19(Suppl 1):42–47. doi: 10.1111/j.1365-2893.2011.01523.x. [DOI] [PubMed] [Google Scholar]
- 136.Neumann-Haefelin C, Thimme R. Adaptive immune responses in hepatitis C virus infection. Curr Top Microbiol Immunol. 2013;369:243–262. doi: 10.1007/978-3-642-27340-7_10. [DOI] [PubMed] [Google Scholar]
- 137.Nishio T, Iimuro Y, Nitta T, Harada N, Yoshida M, Hirose T, et al. Increased expression of collagenase in the liver induces hepatocyte proliferation with cytoplasmic accumulation of beta-catenin in the rat. J Hepatol. 2003;38:468–475. doi: 10.1016/s0168-8278(03)00013-8. [DOI] [PubMed] [Google Scholar]
- 138.Nitta T, Kim JS, Mohuczy D, Behrns KE. Murine cirrhosis induces hepatocyte epithelial mesenchymal transition and alterations in survival signaling pathways. Hepatology. 2008;48:909–919. doi: 10.1002/hep.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–S214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Obokata H, Sasai Y, Niwa H, Kadota M, Andrabi M, Takata N, et al. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature. 2014;505:676–680. doi: 10.1038/nature12969. [DOI] [PubMed] [Google Scholar]
- 141.Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, et al. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014;505:641–647. doi: 10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- 142.Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, et al. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 2010;51:1712–1723. doi: 10.1002/hep.23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Omenetti A, Porrello A, Jung Y, Yang L, Popov Y, Choi SS, et al. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J Clin Invest. 2008;118:3331–3342. doi: 10.1172/JCI35875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ortiz V, Berenquer M, Rayon JM, Carrasco D, Berenquer J. Contribution of obesity to hepatitis C-related fibrosis progression. Am J Gastroenterol. 2002;97:2408–2414. doi: 10.1111/j.1572-0241.2002.05995.x. [DOI] [PubMed] [Google Scholar]
- 145.Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pangburn MK, Ferreira VP, Cortes C. Discrimination between host and pathogens by the complement system. Vaccine. 2008;26(Suppl 8):I15–I21. doi: 10.1016/j.vaccine.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Paradis V, Zalinski S, Chelbi E, Guedi N, Degos F, Vilgrain V, et al. Hepatocellular carcinomas in patients with metabolic syndrome often develop without significant liver fibrosis: a pathological analysis. Hepatology. 2009;49:851–859. doi: 10.1002/hep.22734. [DOI] [PubMed] [Google Scholar]
- 148.Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, et al. Nonstructural 5A protein activates beta-catenin signaling cascades : implication of hepatitis C virus-induced liver pathogenesis. J Hepatol. 2009;51:853–864. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 149.Peng L, Liang D, Tong W, Li J, Yuan Z. Hepatitis C virus NS5A activates the mammalian target of rapamycin (mTOR) pathway, contributing to cell survival by disrupting the interaction between FK506-binding protein 38 (FKBP38) and mTOR. J Biol Chem. 2010;285:20870–20881. doi: 10.1074/jbc.M110.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Philips GM, Chan IS, Swiderska M, Schroder VT, Guy C, Karaca GF, et al. Hedgehog signaling antagonist promotes regression of both liver fibrosis and hepatocellular carcinoma in a murine model of primary liver cancer. PLoS One. 2011;6:e23943. doi: 10.1371/journal.pone.0023943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poveda E, Wyles DL, Mena A, Pedreira JD, Castro-Iglesias A, Cachay E. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 2014;108C:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 152.Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan X, et al. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene. 2014;33:2826–2835. doi: 10.1038/onc.2013.225. [DOI] [PubMed] [Google Scholar]
- 153.Ray R. Progress toward development of a hepatitis C vaccine with broad shoulders. Sci Transl Med. 2011;3:94ps33. doi: 10.1126/scitranslmed.3002772. [DOI] [PubMed] [Google Scholar]
- 154.Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S, et al. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis. 2010;202:862–866. doi: 10.1086/655902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ray RB, Meyer K, Steele R, Shrivastava A, Aggarwal BB, Ray R. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256–2259. doi: 10.1074/jbc.273.4.2256. [DOI] [PubMed] [Google Scholar]
- 156.Ray RB, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 157.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raychoudhury A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, et al. Hepatitis C virus infection impairs IRF-7 translocation and alpha interferon synthesis in immortalized human hepatocytes. J Virol. 2010;84:10991–10998. doi: 10.1128/JVI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Regimbeau JM, Colombat M, Mognol P, Durand F, Abdalla E, Degott C, et al. Obesity and diabetes as a risk factor for hepatocellular carcinoma. Liver Transpl. 2004;10:S69–S73. doi: 10.1002/lt.20033. [DOI] [PubMed] [Google Scholar]
- 160.Roberts AP, Lewis AP, Jopling CL. The role of microRNAs in viral infection. Prog Mol Biol Transl Sci. 2011;102:101–139. doi: 10.1016/B978-0-12-415795-8.00002-7. [DOI] [PubMed] [Google Scholar]
- 161.Roingeard P. Hepatitis C virus diversity and hepatic steatosis. J Viral Hepat. 2013;20:77–84. doi: 10.1111/jvh.12035. [DOI] [PubMed] [Google Scholar]
- 162.Rouille Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, et al. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Saito K, Ait-Goughoulte M, Truscott SM, Meyer K, Blazevic A, Abate G, et al. Hepatitis C virus inhibits cell surface expression of HLA-DR, prevents dendritic cell maturation, and induces interleukin-10 production. J Virol. 2008;82:3320–3328. doi: 10.1128/JVI.02547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Saito K, Meyer K, Warner R, Basu A, Ray RB, Ray R. Hepatitis C virus core protein inhibits tumor necrosis factor alpha-mediated apoptosis by a protective effect involving cellular FLICE inhibitory protein. J Virol. 2006;80:4372–4379. doi: 10.1128/JVI.80.9.4372-4379.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virunonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Scarselli E, AnsuiniH, Cerino R, Roccasecca M, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schulze-Krebs A, Preimel D, Popov Y, Bartenschlager R, Lohmann V, Pinzani M, et al. Hepatitis C virus-replicating hepatocytes induce fibrogenic activation of hepatic stellate cells. Gastroenterology. 2005;129:246–258. doi: 10.1053/j.gastro.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 168.Seitz HK, Stickel F. Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem. 2006;387:349–360. doi: 10.1515/BC.2006.047. [DOI] [PubMed] [Google Scholar]
- 169.Sheikh MY, Choi J, Qadri I, Friedman JE, Sanyal AJ. Hepatitis C virus infection: molecular pathways to metabolic syndrome. Hepatology. 2008;47:2127–2133. doi: 10.1002/hep.22269. [DOI] [PubMed] [Google Scholar]
- 170.Sherman M. Hepatocellular carcinoma: epidemiology, surveillance and diagnosis. Semin Liver Dis. 2010;30:3–16. doi: 10.1055/s-0030-1247128. [DOI] [PubMed] [Google Scholar]
- 171.Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, et al. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37:138–145. doi: 10.1038/emm.2005.19. [DOI] [PubMed] [Google Scholar]
- 172.Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, et al. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840–848. doi: 10.1053/j.gastro.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 173.Shrivastava S, Mukherjee A, Ray R, Ray RB. Hepatitis C virus induces interleukin-1ß/IL-18 in circulatory and resident liver marcrophages. J Virol. 2013;87:12284–12290. doi: 10.1128/JVI.01962-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. World J Hepatol. 2013;5:479–486. doi: 10.4254/wjh.v5.i9.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shrivastava S, Ray RB. Hepatitis C virus infection, autophagy and innate immune response. In: Hayat MA, editor. Autophagy. Cancer, other pathologies, inflammation, immunity, infection and aging. Vol. 3. Amsterdam: Academic Press, Elsevier; 2014. pp. 164–190. [Google Scholar]
- 176.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L, et al. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478–1488. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tang Y, Kitjsin K, Jogunoori W, Li C, Deng CX, Muller SC, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;19:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 181.Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, et al. Hepatitis C virus core protein upregulates transforming growth factor-beta1 transcription. J Med Virol. 2004;72:52–59. doi: 10.1002/jmv.10545. [DOI] [PubMed] [Google Scholar]
- 182.Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. Hepatitis C virus polyprotein processing : kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J Virol. 1994;68:8418–8422. doi: 10.1128/jvi.68.12.8418-8422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Taura K, De Minicis S, Seki E, Hatano E, Iwaisako K, Osterreicher CH, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135:1729–1738. doi: 10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 184.Theret N, Musso O, Turlin B, Lotrian D, Bioulac-Sage P, Campion JP, et al. Increased extracellular matrix remodeling is associated with tumor progression in human hepatocellular carcinomas. Hepatology. 2001;34:82–88. doi: 10.1053/jhep.2001.25758. [DOI] [PubMed] [Google Scholar]
- 185.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 186.Todorovic V, Rifkin DB. LTBPs, more than just an escort service. J Cell Biochem. 2012;113:410–418. doi: 10.1002/jcb.23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Torre D, Zeroli C, Giola M, Ferrario G, Fiori GP, Bonetta G, et al. Serum levels of interleukin-1 alpha, interleukin-1 beta, interleukin-6, and tumor necrosis factor in patients with acute viral hepatitis. Clin Infect Dis. 1994;18:194–198. doi: 10.1093/clinids/18.2.194. [DOI] [PubMed] [Google Scholar]
- 188.van Zijl F, Mair M, Csiszar A, Schneller D, Zulehner G, Huber H, et al. Hepatic tumor-stroma crosstalk guides epithelial to mesenchymal transition at the tumor edge. Oncogene. 2009;28:4022–4033. doi: 10.1038/onc.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Veerapu NS, Park SH, Tully DC, Allen TM, Rehermann B. Trace amounts of sporadically reappearing HCV RNA can cause infection. J Clin Invest. 2014;124:3469–3478. doi: 10.1172/JCI73104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132:667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 191.Wang QM, Hockman MA, Staschke K, Johnson RB, Case KA, Lu J, et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology. 2008;47:1394–1400. doi: 10.1002/hep.22193. [DOI] [PubMed] [Google Scholar]
- 193.Williams P, Chaudhry Y, Goodfellow IG, Bilington J, Powell R, Spiller OB, et al. Mapping CD55 function. The structure of two pathogen-binding domains at 1.7A. J Biol Chem. 2003;278:10691–10696. doi: 10.1074/jbc.M212561200. [DOI] [PubMed] [Google Scholar]
- 194.Wilson GK, Brimacombe CL, Rowe IA, Reynolds GM, Fletcher NF, Stamataki Z, et al. A dual role for hypoxia inducible factor-1α in the hepatitis C virus life cycle and hepatoma migration. J Hepatol. 2012;56:803–809. doi: 10.1016/j.jhep.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wu Y, Zuo J, Ji G, Saiyin H, Liu X, Yin F, et al. Proapoptotic function of integrin beta(3) in human hepatocellular carcinoma cells. Clin Cancer Res. 2009;15:60–69. doi: 10.1158/1078-0432.CCR-08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yaqoob U, Cao S, Shergill U, Jagavelu K, Geng Z, Yin M, et al. Neuropilin-1 stimulates tumor growth by increasing fibronectin fibril assembly in the tumor microenvironment. Cancer Res. 2012;72:4047–4059. doi: 10.1158/0008-5472.CAN-11-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yasui K, Hashimoto E, Komorizono Y, Koike K, Arii S, Imai Y, et al. Characteristics of patients with nonalcoholic steatohepatitis who develop hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2011;9:428–433. doi: 10.1016/j.cgh.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 198.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signaling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 199.Yin M, Talwalkar JA, Glaser KJ, Manduca A, Grimm RC, Rossman PJ, et al. Assessment of hepatic fibrosis with magnetic resonance elastography. Clin Gastroenterol Hepatol. 2007;5:1207–1213.e2. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zeisberg M, Yang C, Martino M, Duncan MB, Rieder F, Tanjore H, et al. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–23347. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 201.Zekri AR, Bahnassy AA, Abdel-Wahab SA, Khafagy MM, Loutfy SA, Radwan H, et al. Expression of pro-and anti-inflammatory cytokines in relation to apoptotic genes in Egyptian liver disease patients associated with HCV-genotype-4. J Gastroenterol Hepatol. 2009;24:416–428. doi: 10.1111/j.1440-1746.2008.05699.x. [DOI] [PubMed] [Google Scholar]
- 202.Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56:769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang H, Ozaki I, Mizuta T, Matsuhashi S, Yoshimura T, Hisatomi A, et al. Beta 1-integrin protects hepatoma cells from chemotherapy induced apoptosis via a mitogen-activated protein kinase dependent pathway. Cancer. 2002;95:896–906. doi: 10.1002/cncr.10751. [DOI] [PubMed] [Google Scholar]
- 204.Zhou X, Jamil A, Nash A, Chan J, Trim N, Iredale JP, et al. Impaired proteolysis of collagen I inhibits proliferation of hepatic stellate cells: implications for regulation of liver fibrosis. J Biol Chem. 2006;281:39757–39765. doi: 10.1074/jbc.M605621200. [DOI] [PubMed] [Google Scholar]
- 205.Zimmermann HW, Trautwein C, Tacke F. Functional role of monocytes and macrophages for the inflammatory response in acute liver injury. Front Physiol. 2012;3:56. doi: 10.3389/fphys.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]


